Early introduction of appropriate antibiotherapy is one of the major prognostic-modifying factors in community acquired pneumonia (CAP). Despite established guidelines for empirical therapy, several factors may influence etiology and, consequently, antibiotic choices. The aims of this study were to analyze the etiology of CAP in adults admitted to a northern Portugal University Hospital and evaluate the yield of the different methods used to reach an etiological diagnosis, as well as analyze of the impact of patient demographic and clinical features on CAP etiology.
We retrospectively analyzed 1901 cases of CAP with hospitalization. The diagnostic performance increased significantly when blood and sputum cultures were combined with urinary antigen tests. The most frequent etiological agent was Streptococcus pneumoniae (45.7%), except in August, when it was overtaken by gram-negative bacilli (GNB) and Legionella pneumophila infections. Viral infections were almost exclusive to winter and spring. A negative microbiological result was associated with increasing age, non-smoking and lack of both blood/sputum cultures. Younger age was a predictor for S. pneumoniae, Influenza and L. pneumophila infections. Active smoking without any previously known respiratory disease was a risk factor for legionellosis. COPD was associated with Haemophilus influenzae cases, while dementia was typical in GNB and S. aureus patients. Diabetes mellitus (DM) and heart disease were negative predictors of S. pneumoniae and H. influenzae, respectively. P. aeruginosa was an independent risk factor for mortality (OR 13.02, 95% CI 2.94–57.7).
This study highlights the importance of a comprehensive microbiological diagnostic workup and provides clues to predicting the most probable CAP causative agents, based on a patient's clinical profile. These may be taken into account when establishing first line antibiotherapy.
Community-acquired pneumonia (CAP) remains a major cause of morbidity and mortality cause in Western countries, despite significant advances in antimicrobial and supportive therapy.1 In Portugal, disease burden has been increasing in the last decade, especially among the elderly. According to the latest data, pneumonia is the main cause of respiratory hospitalization, accounting for 3.7% of all hospital admissions in Portugal between 2000 and 2009, with an intra-hospital mortality rate of 20.4%.2
Early introduction of appropriate therapy figures as one of the major prognostic modifying factors.3 The choice of initial antimicrobial treatment is usually empirical and it is essential that it is broad enough to cover all the likely pathogens. Despite international guidelines recommending specific empirical regimens, local differences in etiological pathogens have been found.4 More recently, the emergence of antibiotic-resistant agents has created new concerns for a more judicious use of broad-spectrum antibiotics, which poses important challenges in initiating early therapy.5 For this reason, and in order to propose the most appropriate treatment in a specific context, it is crucial to establish microbiological epidemiology data within different healthcare settings.
Respiratory bacteria are the major causative microorganisms, Streptococcus pneumoniae being the most common etiological CAP agent.4 Other frequently identified pathogens are Haemophilus influenzae, Gram-negative enteric bacilli and respiratory viruses.6 However, certain clinical features of patients have been associated with the risk of infection by multidrug-resistant (MDR) pathogens.7 Some of these cases have been designated as healthcare-associated pneumonia (HCAP), which includes patients living in nursing homes, who are immunosuppressed, had recent hospitalization, undergo chronic dialysis or home-based infusion therapy. This differentiation was supported by the contrasting observed outcomes, with higher mortality rates among those with pneumonia acquired in the hospital or associated with healthcare environments.8 However, this is a controversial term, given that a few European studies have shown etiology patterns in HCAP patients similar to those found in CAP patients,9 which contrasts with data from the USA.9 Moreover, etiological agents may have a seasonal distribution.10 These variables must be taken into account when approaching the patient diagnosed with pneumonia.
The primary aim of this study was to describe the etiology of CAP in adults admitted to a northern Portugal University Hospital. Secondary aims are the evaluation of the yield of the diagnostic methods applied as well as the analysis of the impact of patients’ demographic and clinical features on CAP microbiological etiology.
MethodsStudy designRetrospective analysis of hospitalized adults due to CAP at a northern Portugal University Hospital from January 2013 to December 2015. CAP was diagnosed in accordance with the IDSA/ATS guidelines as the presence of (i) at least one of the clinical symptoms of cough, sputum, fever, dyspnea, and pleuritic chest pain, (ii) elevated inflammatory biomarkers and (iii) new or evolving pulmonary infiltrate on chest radiography.11 These criteria had to be present within 48h of admission. Exclusion criteria were: age under 18 years; active tuberculosis; noninfectious diseases such as pulmonary infarction and pulmonary edema; hospital-acquired pneumonia (occurring ≥48h after admission); or patients presenting features that were historically associated with HCAP (hospitalization for ≥2 days in the 90 days before admission, outpatient infusion therapy or chemotherapy, home wound care in the previous 30 days, admission from a nursing home or long-term care facility, or chronic dialysis in a hospital or clinic).7
The study was approved by the Health Ethics Committee of the hospital (approval number 2019-CE-P002). The requirement to obtain informed written consent from each individual was waived, as the study was limited to the review of existing medical records. To ensure confidentiality, each case was anonymized by the assignment of a random identification number.
Microbial sample collection and analysisBefore commencing antibiotic therapy, the common practice in the hospital is to obtain from every patient hospitalized with CAP at least two blood cultures (BD BACTEC FX™ Blood Culture System, Sparks, Maryland, USA), urine sample for specific antigen detection and, whenever possible, sputum specimens for Gram stain and culture. During flu season, recommendations also included performance of nasopharyngeal swab based on clinical suspicion. Bronchoalveolar lavage (BAL) and diagnostic thoracentesis were performed on medical indication, based on clinical judgement. Serological investigation was not routinely done. Bacteriological specimens were cultured on standard media. Sputum samples were considered acceptable for culture only when displaying >25 polymorphonuclear leukocytes and <10 squamous epithelial cells per 100× power field.12 Most agents – S. aureus, S. agalactiae, S. hominis, Enterobacterales, Pseudomonas aeruginosa, Acinetobacter baumannii and Stenotrophomonas maltophilia – were identified by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry using the VITEK® MS analytical system, or VITEK cards (bioMérieux, France). For S. pneumoniae identification the test of susceptibility to the optochin were used, with bile solubility as a confirmatory test. Antimicrobial susceptibility was accessed mainly through the Kirby–Bauer method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) clinical breakpoints, or by VITEK® cards. Urinary antigen detection tests for S. pneumoniae and Legionella pneumophila serogroup 1 were executed with the BinaxNOW pneumococcal or Legionella urinary antigen test (Binax, ME, USA). Nasopharyngeal swab was analyzed by real-time reverse-transcription PCR (RT-PCR) for detection of RNA viruses, namely influenza A and B viruses, and H1N1 on influenza A virus positive samples, parainfluenza viruses types 1–3, metapneumovirus, rhinovirus, enterovirus and respiratory syncytial virus. Real-time PCR detection of L. pneumophila and/or Pneumocystis jirovecii in respiratory samples was performed when medically relevant.
Statistical analysisAll the statistical analyses were performed using the GraphPad Prism® 7 software (GraphPad Software, Inc.) and SPSS software program, version 25 (IBM® SPSS®, Inc.). Continuous variables were expressed as mean and standard deviation (SD), whereas categorical variables were expressed as frequency, unless stated otherwise. Frequency comparison was done via the χ2 test or the Fisher exact test for the categorical variables and the Student t-test or the Mann–Whitney U test for the continuous variables.
Different regression logistic multivariate models were performed separately to predict each microbiological etiology as the dependent variable. The Hosmer–Lemeshow goodness-of-fit test was performed to assess the overall fit of the model. A P-value <0.05 was considered significant for all analyses.
ResultsDemographics and clinical backgroundAfter applying the selection criteria, the final study population consisted of 1901 patients. Clinico-demographic characteristics and initial antimicrobial treatments are presented in Table 1. Over half (55.7%) of the CAP patients were male, and mostly older people, 71.1% of the study population were ≥65 years old. Most patients (73.3%) had at least one comorbid condition and 30.2% had ≥2 comorbidities (data not shown). Previous respiratory disease was common (26.4%) with chronic obstructive pulmonary disease (COPD) accounting for the majority of cases (18.8%). Non-respiratory conditions were also considerably prevalent with diabetes mellitus (DM) and heart disease being present in 418 and 401 patients (22% and 21.1%, respectively).
Demographics and clinical background of the 1901 hospitalized adult patients with community-acquired pneumonia.
| Characteristics | No. of patients (%)a |
|---|---|
| Age (years), mean±SD | 72.0±16.5 |
| 18–39 years | 100 (5.3) |
| 40–64 years | 449 (23.6) |
| 65–79 years | 581 (30.6) |
| ≥80 years | 771 (40.6) |
| Male/female gender | 1058 (55.7)/843 (44.3) |
| Active smoker | 338 (19.6)b |
| Comorbid conditions | |
| Alcohol abuse | 85 (4.5) |
| Diabetes mellitus | 418 (22) |
| Heart disease | 401 (21.1) |
| Chronic kidney disease | 211 (11.1) |
| Asthma | 28 (1.5) |
| COPD | 357 (18.8) |
| Structural lung diseasec | 117 (6.2) |
| Immunosuppressiond | 80 (4.2) |
| HIV infection | 121 (6.4) |
| Active malignancy | 133 (7.0) |
| Dementia | 199 (10.5) |
| Antimicrobial treatments,n(%) | |
| Monotherapy | |
| Penicillin G | 11 (0.6) |
| Amoxicillin/clavulanate | 289 (15.2) |
| Levofloxacin | 339 (17.8) |
| 3rd gen. cephalosporin | 68 (3.6) |
| Macrolide | 2 (0.1) |
| Piperacillin/tazobactam | 95 (5) |
| Carbapenem | 10 (0.5) |
| Combination providing coverage of ‘atypical’ pathogens | |
| Amox/clav+macrolide | 677 (35.6) |
| 3rd gen. CS+macrolide | 290 (15.3) |
| Pip/taz+macrolide | 58 (3.1) |
| Carbapenem+macrolide | 13 (0.7) |
| Oseltamivir | 11 (0.6) |
| None/unknown | 38 (2) |
Microbiological tests obtained included the following: blood cultures (n=1403, 73.8%), sputum samples culture (n=1058, 55.7%), urinary antigen assays (n=1066, 56.1%), pleural fluid (n=41, 2.2%), BAL (n=6, 0.3%). In 266 cases (14%) no microbiology testing was performed. Out of the remaining patients, those with at least one sample collected, etiological diagnosis was obtained for 420 (25.7%) patients. S. pneumoniae was the most commonly detected agent (45.7%, n=192), followed by Haemophilus influenza (19.8%, n=83), GNB (10%, n=42) – Klebsiella pneumoniae (n=13), Escherichia coli (n=10), Pseudomonas aeruginosa (n=8), Enterobacter spp (n=3), Proteus mirabilis (n=2), Serratia marcescens (n=2), Acinetobacter baumannii (n=2), Raoultella ornithinolytica (n=1), Stenotrophomonas maltophilia (n=1) – Influenza virus (9.0%, n=38), L. pneumophila (7.6%, n=32), S. aureus (4.3%, n=18), Moraxella catarrhalis (1.7%, n=7) and to a lesser extent other bacteria and virus both representing 0.9% (n=4), Table 2.
Etiological findings and contribution of different methods to diagnostic yield in the study population.
| Pathogen | No. (%) of patients with positive findingsa (n=420) | Sputum or BAL samples for culture and/or L. pneumophila or virus PCR (n=327) | Blood culture (n=366) | Pleural fluid culture (n=11) | Urinary antigen test (n=301) | NP/OP swab (n=34) |
|---|---|---|---|---|---|---|
| S. pneumoniae | 192 (45.7) | 46 (14.1) | 65 (17.8) | 2 (18.2) | 134 (44.5) | NA |
| H. influenzae | 83 (19.8) | 70 (21.4) | 15 (4.1) | – | NA | NA |
| GNBb | 42 (10) | 32 (9.8) | 16 (4.4) | 1 (9.1) | NA | NA |
| Influenza virus | 38c (9.0) | 4 (1.2) | – | NA | NA | 31 (91.2) |
| L. pneumophila | 32 (7.6) | 4 (1.2) | – | NA | 30 (10.0) | NA |
| S. aureus | 18 (4.3) | 16 (4.9) | 4 (1.1) | – | NA | NA |
| M. catarrhalis | 7 (1.7) | 7 (2.1) | – | – | NA | NA |
| Other bacteriad | 4 (0.9) | – | 4 (1.1) | – | NA | NA |
| Other viruse | 4 (0.9) | 1 (0.3) | – | – | NA | 3 (8.8) |
Data are number of patients and proportion (%) of cases whose infections were etiologically established by use of a particular method listed. BAL, bronchoalveolar lavage; GNB, gram-negative bacilli; MSSA, methicillin-susceptible Staphylococcus aureus; MRSA, methicillin-resistant S. aureus; NP, nasopharynx; OP, oropharynx; NA, not applicable.
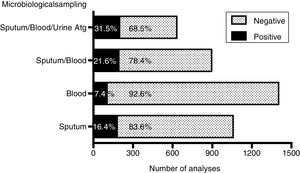
The overall diagnostic yield of blood or sputum cultures was low, with 7.4% (104 positive results in 1403 tests) and 16.4% (174 positive results in 1058 tests), respectively. However, the diagnostic performance increased significantly (P<0.0001) to 21.6% when blood and sputum were both collected, and to 31.5% when urinary antigen tests for S. pneumoniae and L. pneumophila were added to blood and sputum cultures (Fig. 1). The most frequently isolated agent in respiratory samples was H. influenzae (21.4%), followed by S. pneumoniae (14.1%). In fact, there was a significant contribution of blood cultures in the etiological diagnosis of S. pneumoniae, as shown in Table 2, highlighting the importance of blood cultures to isolate the causative pathogen. A negative microbiological result was significantly associated to older patients, female gender, non-smokers, heart disease, chronic kidney disease (CKD), dementia and incomplete etiological diagnosis workup, and correlated negatively with HIV infection. Three independent predictors for unknown etiology were identified in multivariate analysis: increasing years of age (P<0.001), non-smoking (P=0.025) and lack of both blood/sputum cultures (P<0.001) (Table 3).
Causal microorganisms depending on clinical background and independent risk factors for each etiological agent.
| Unknown etiology (n=1481) | S. pneumoniae (n=192) | H. influenzae (n=83) | GNB (n=42) | Influenza virus (n=38) | L. pneumophila (n=32) | S. aureus (n=18) | M. catarrhalis (n=7) | |
|---|---|---|---|---|---|---|---|---|
| Univariate analysis | ||||||||
| Age (years), mean±SD | 73.3±16.0***H | 68.7±16.4**L | 69.4±17.3 | 74.1±16.1 | 54.8±14.9***L | 58.1±16.5***L | 63.8±20.3 | 79.6±7.1 |
| Male gender, n (%) | 806 (54.4)*L | 104 (75.2) | 55 (66.3)*H | 24 (57.1) | 21 (55.3) | 25 (78.1)*H | 12 (66.7) | 4 (57.1) |
| Winter & Spring, n (%) | 1007 (68.0) | 136 (70.8) | 70 (84.3)**H | 25 (59.5) | 37 (97.4)***H | 8 (25)***L | 16 (88.9) | 7 (100) |
| Active smoker, n (%)a | 228 (15.4)***L | 45 (23.4)*H | 23 (27.7)*H | 6 (14.3) | 14 (36.8)*H | 19 (59.4)***H | 1 (5.6) | 0 |
| Comorbid conditions, n (%) | ||||||||
| Alcohol abuse | 63 (4.3) | 13 (6.8) | 4 (4.8) | 2 (4.8) | 0 | 3 (9.4) | 0 | 0 |
| Diabetes mellitus | 337 (22.8) | 29 (15.1)*L | 18 (21.7) | 9 (21.4) | 11 (28.9) | 8 (25) | 4 (22.2) | 0 |
| Heart disease | 333 (22.5)**H | 35 (18.2) | 11 (13.3)*L | 7 (16.7) | 4 (10.5) | 4 (12.5) | 2 (11.1) | 3 (42.9) |
| Chronic kidney disease | 178 (12.0)*H | 12 (6.3)*L | 9 (10.8) | 7 (27.5) | 2 (5.3) | 1 (3.1) | 2 (11.1) | 0 |
| Asthma | 21 (1.4) | 5 (2.6) | 0 | 0 | 2 (5.3) | 0 | 0 | 0 |
| COPD | 268 (18.1) | 41 (21.4) | 28 (33.7)**H | 6 (14.3) | 8 (21.1) | 2 (6.3) | 0*L | 3 (42.9) |
| Structural lung diseaseb | 94 (6.3) | 11 (5.7) | 3 (3.6) | 2 (4.8) | 4 (10.5) | 0 | 2 (11.1) | 0 |
| Immunosuppressionc | 64 (4.3) | 6 (3.1) | 2 (2.4) | 1 (2.4) | 4 (10.5) | 1 (3.1) | 1 (5.6) | 0 |
| HIV infection | 84 (5.7)*L | 19 (9.9) | 10 (12.0)*H | 2 (4.8) | 1 (2.6) | 2 (6.3) | 1 (5.6) | 0 |
| Active malignancy | 109 (7.4) | 14 (7.3) | 4 (4.8) | 3 (7.1) | 1 (2.6) | 1 (3.1) | 1 (5.6) | 0 |
| Dementia | 170 (11.5)**H | 13 (6.8) | 3 (3.6)*L | 8 (19.0) | 0*L | 1 (3.1) | 4 (22.2) | 0 |
| Both blood and sputum cultures, n (%) | 609 (41.1)***L | 117 (60.9)***H | 62 (74.7)***H | 28 (66.7)**H | 32 (84.2)***H | 17 (53.1) | 16 (88.9)***H | 6 (85.7)*H |
| Death, n (%) | 173 (11.7) | 19 (9.9) | 5 (6.0) | 13 (31.0)***H | 5 (13.1) | 4 (12.5) | 3 (16.7) | 0 |
| Multivariate regression analysis | ||||||||
| Predictor, OR (95% CI), P-value | Age,1.01 (1.01–1.02), <0.001Active smoker,0.72 (0.53–0.96), 0.025Blood & Sputum culture,0.34 (0.26–0.43), <0.001 | Age,0.99 (0.98–0.99), 0.025Diabetes mellitus,0.63 (0.41–0.95), 0.027Blood & Sputum culture,1.80 (1.32–2.45), <0.001 | Winter & Spring,2.23 (1.22–4.08), 0.01Heart disease,0.52 (0.27–0.99), 0.05COPD,2.12 (1.31–3.43), 0.002Blood & Sputum culture,3.22 (1.94–5.34), <0.001 | Dementia,2.26 (1.03–4.99), 0.043Blood & Sputum culture,2.39 (1.25–4.59), 0.009 | Age,0.95 (0.93–0.97), <0.001Winter & Spring,14.01 (1.91–102.99), 0.009Blood & Sputum culture,4.71 (1.94–11.44), 0.001 | Age,0.97 (0.95–0.99), 0.021Winter & Spring,0.14 (0.06–0.34), <0.001Active smoker,5.58 (2.28–13. 61), <0.001Any respiratory disease,0.18 (0.04–0.76), 0.020 | Dementia,3.01 (0.98–9.46), 0.054Blood & Sputum culture,9.80 (2.24–42.93), 0.002 | |
Data represent number (percentage) of patients for each variable, except patient age, which is presented as mean±SD. Each percentage was compared (Pearson's chi-square or Fisher's exact tests) to that of patients without the condition (data not shown). *L/*H Significantly lower/higher (respectively) than in patients without the condition (*P<0.05, **P<0.01, ***P<0.001).
Bronchiectasis, pleuritis or sequelae of prior tuberculosis conditioning alteration of the normal architecture of the lung.
Immunosuppressive drugs were defined as any use of systemic steroids, azathioprine, mycophenolate mofetil, TNF-alpha inhibitor, Cyclosporine, Cyclophosphamide and/or Methotrexate within previous 3 months. CI, confidence interval. COPD, chronic obstructive pulmonary disease. HIV, human immunodeficiency virus. OR, odds ratio.
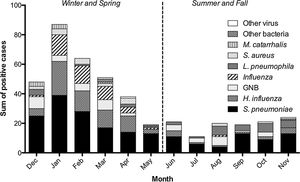
Winter and spring accounted for the majority of adult CAP cases, with highest prevalence of S. pneumoniae, H. influenzae and Influenza virus during the coldest half of the year. In all 3 years assessed, the peak month of hospitalization was January, to which the annual outbreaks of flu largely contributed. In fact, when compared to bacterial CAP cases, viral infections were almost exclusive to the winter and spring (Fig. 2). S. pneumoniae was the most frequently identified etiological agent in all months, except in August, when it was overtaken by GNB and L. pneumophila. In the multivariate analysis, winter and spring season was a positive predictor for H. influenzae (OR 2.23, 95% CI 1.22–4.08, P=0.01) and Influenza virus (OR 14.01, 95% CI 1.91–102.99, P=0.001), and a negative one for L. pneumophila (OR 0.14, 95% CI 0.06–0.34, P=0.001) (Table 3).
CAP etiology according to patients’ characteristicsThe distribution of causal microorganisms varied depending on age and comorbidities (Table 3). Multivariate logistic regression analyses were performed to determine the independent risk factors for each etiological agent. Increasing age was a negative predictor for S. pneumoniae, Influenza and L. pneumophila. Diabetes and heart disease were significantly less common in patients with CAP due to S. pneumoniae (OR 0.63, 95% CI 0.41–0.95, P=0.027) and H. influenzae (OR 0.52, 95% CI 0.27–0.99, P=0.05), respectively. Active smoking (OR 5.58, 95% CI 2.28–13.61, P<0.001) and absence of any respiratory disease (OR 0.18, 95% CI 0.04–0.76, P=0.02) were risks factors for legionellosis. Conversely, COPD was an important risk factor for H. influenzae cases (OR 2.12, 95% CI 1.31–3.43, P=0.002). Dementia was associated with increased risk for GNB (OR 2.26, 95% CI 1.03–4.99, P=0.043) and S. aureus (OR 3.01, 95% CI 0.98–9.46, P=0.054). Blood/sputum cultures were used as a constant covariate across groups, to normalize data for those with most complete microbiological investigation. The exception to this rule was L. pneumophila, as this parameter had no significant impact, since almost all diagnoses were made using urinary antigen test.
Mortality ratesThe in-hospital mortality rate was 11.7%. Fifty of the 223 patients who died had etiological diagnosis and GNB were the most prevalent agents among the deceased. Within this group, this tendency was pushed by the 5 fatal cases out of 8 (62.5%) infected with Pseudomonas aeruginosa (P<0.001). Bacteremia was also related with higher mortality, with 17 (16.3%) fatalities among 104 positive blood cultures, when compared to negative tests (135/1299, 10.4%; P=0.06). Multiple logistic regression analysis confirmed P. aeruginosa infection as an independent mortality risk factor (OR 13.02, 95% CI 2.94–57.7, P=0.001).
DiscussionThis study provides a comprehensive insight into the etiological and clinical profile of CAP hospitalized patients in Portugal and describes the real-life microbiological testing in this setting. Proper diagnostic testing improves clinical outcomes directly through individualization of antibiotic management, and indirectly by delivering relevant epidemiological data that influences initial empirical therapy. Although a fundamental exercise, establishing microbiological CAP diagnosis is challenging.4 In a prospective study, when conventional methods (i.e., bacterial cultures, urinary antigen assays, serology) were combined with PCR-based methods, definitive or probable etiology was established in 63% of cases.13 In our analysis, with complete standard microbiological testing (sputum, blood culture and urine antigen test), we attained a microbial confirmation in 31.5% of cases. While it represents a lower diagnostic rate than the described in prospective studies,14–16 it is comparable to previous large real-life studies.17,18 Due to the retrospective nature of this work, harvesting of all products was not possible in some cases, and 14% of the patients in the cohort did not undergo any microbiology test. We may speculate that clinicians are less prone to undergo invasive microbiological assessment in patients with the poorest performance status. Indeed, patients without microbiological studies were significantly older.
Additionally, our work confirms the low sensitivity of blood cultures described in other reports18,19 with only 7.4% positive results. The low yield of blood cultures could be possibly explained by delay in sample collection and/or prior use of antibiotics, but these findings must also depend on bacterial infection biology, as there was a higher rate of bacteremia among pneumococcal disease. Microbial yield of sputum culture was also very modest. The poor quality of sputum accounted for several missing diagnoses (data not shown), further aggravated by ineffective cough or inappropriate sputum production, particularly at the older population fringe. Finally, once serologic assays are not routinely performed in our center, Mycoplasma and Chlamydophila pneumoniae, and supposedly several other cases of viral infections, could not be detected, which also affected importantly the overall diagnosis rate. Many studies conducted prospectively, performing serum IgM antibody measurement, combined or not with PCR detection, described atypical bacteria as major causes of CAP, with variable frequencies up to 18%, and the viruses involved in up to 30% of hospitalized CAP, only surpassed by S. pneumoniae as the most frequent agents.20 In the present work, viral etiology was probably underestimated, having been established by PCR of samples collected by pharyngeal swabs in selected patients, based on clinical judgement. Nevertheless, it is acknowledged that serology testing has little impact on the routine management of the individual patient.20,21 Bearing this in mind and considering the high healthcare costs associated, there is a general notion that serologies are more useful in epidemiological studies than in clinical practice.
Considering the low microbial detection yield of various works, some authors have claimed that current empirical antimicrobial recommendations are based on weak evidence.18 Knowing the importance of the early implementation of appropriate therapy, we derived etiological predictors from CAP patients’ features, to better suit therapy to the most probable causative agent. Our data suggest that, in older people, S. pneumoniae, Influenza and L. pneumophila are less common, and a higher incidence of CAP is observed caused by GNB, S. aureus and M. catarrhalis, pathogens that more frequently present drug resistances. This is important, since CAP is a growing problem among the elderly.1,22
Globally, S. pneumoniae remains the most common detected cause of CAP4 and our results confirm these data, with 45.7% of the identified cases. However, there was a large contribution of the urine antigen test for pneumococcal diagnoses, when a similar method is not routinely available to other microbial agents. In fact, if we consider etiological diagnosis based on respiratory samples alone, we find H. influenza as the most commonly isolated organism. It is known that H. influenzae is a common CAP pathogen in older patients and those with respiratory comorbidities, such as COPD, as is the case in our sample population. These patients usually present chronic bronchitis and are frequent sputum producers, which facilitates the noninvasive collection of appropriate sputum samples improving the diagnostic yield these cultures.
Both S. pneumoniae and H. influenzae may exist as commensal organisms of the upper respiratory tract, so quantitative multiplex nucleic acid amplification test (NAAT) would detect and differentiate the etiologic agent of CAP better. A study from the United Kingdom identified an etiologic agent by quantitative PCR in 87% of CAP patients, including S. pneumoniae in 36% and H. influenzae in 40%.23
Future studies are needed to unravel how demographic changes in European countries will impact current microbial epidemiology. Furthermore, it is expected that the incidence of CAP by S. pneumoniae will decrease due to the introduction of pneumococcal vaccines.24 Thus, upcoming empirical antimicrobial treatment recommendations may have to take into special account the rise of beta-lactamase-producing agents in this setting, including H. influenza, M. catarrhalis and GNB. We report a significant proportion of CAP cases due to GNB (∼10%), with Klebsiella pneumoniae and P. aeruginosa comprising nearly half of these cases, which is only paralleled in a few other reports.25,26 The highest incidence occurred in patients suffering from dementia and were associated with increased mortality of up to 30%. When controlling for age, dementia and other disabling comorbidities, P. aeruginosa, but not GNB as a whole, was an independent risk factor for mortality. These findings are identical to Spanish data,25 where the authors reported 11% of GNB infections, associated with 32% mortality, much higher than observed with the non-GNB group (9%). In that publication, however, P. aeruginosa failed to be an independent predictor of death.25 Similar effect on mortality related to GNB was found in Asian countries27 and South America.28 Unfortunately, there are no specific measures to prevent pneumonia caused by GNB in the community setting. Despite this, since dementia was an independent risk for GNB infections in our study, measures promoting good oral hygiene and minimizing the use of proton-pump inhibitors can help to reduce bacterial microaspiration among these patients. Additionally, hand feeding should be tried before considering tube feeding and a semi-recumbent position with the head of the bed at a 30–45° angle should be adopted in bedridden patients. Another well-recognized risk factor for GNB infection or other drug-resistant bacteria is previous antibiotic treatment. In line with this, we believe that our results may contribute to antibiotic stewardship and decrease the prevalence of multidrug-resistant organisms.
Moreover, one of the highlights of the paper was the seasonal variability of microbiological agents. Seasonality was particularly important for L. pneumophila infections that occurred almost exclusively during summer and Fall. Given the potential severity of the disease, we propose that clinicians should have higher suspicion index and undergo more intensive diagnostic workup to exclude Legionellosis during that period. A similar approach has been already taken during Influenza season with active virus search during winter, with impact on treatment choices.
Some limitations of the study must be acknowledged. This was a retrospective observational study that could not evaluate all the clinical parameters that may affect etiology. Moreover, a complete microbiological evaluation was not performed in all patients and failure to perform serology cause potential bias in agent identification. Finally, this work was conducted in a particular setting, only including hospitalized patients in a tertiary northern Portuguese hospital. Therefore, these data may not reflect cases of less severity, diagnosed at small community hospitals or in other geographical regions.
In conclusion, besides promoting smoking cessation and encouraging flu and anti-pneumococcal vaccination, investing in the etiological diagnosis, taking into account the most probable agents in a given clinical context can lead to better antibiotic selection thus improving outcomes. Our data suggest that initial empirical coverage of GNB, including P. aeruginosa, should be considered in the elderly with dementia, since these patients usually have a worse prognosis and fail to provide appropriate sputum samples for culture.
Further studies on the epidemiology and prognostic risk factors for CAP caused by GNB and other resistance-prone agents are warranted to assess if early recognition of microbial etiology can modify the outcomes.
Conflict of interestsThe authors declare no conflict of interests.