Substantial progress has been made over the last years in understanding critical molecular and cellular mechanisms driving tumor initiation and progression, with more than 50% of lung adenocarcinomas the main subtype of non-small-cell lung cancer (NSCLC) harboring oncogenic drivers.1–3 These findings led to the development of several novel drugs and treatment strategies and shifted the treatment paradigm of advanced NSCLC from a morphology-based to a predictive biomarker-driven approach based on tumor molecular genotyping.
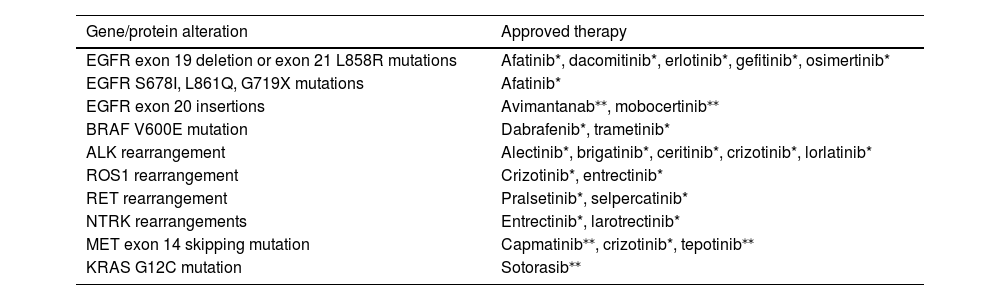
Targeted therapies are the first treatment option for patients with advanced or metastatic disease with tumors harboring oncogenic mutations. In the absence of targetable oncogenic drivers, immunotherapy, either in monotherapy or combination, is the treatment of choice. Molecular alterations with approved therapies in NSCLC are depicted in Table 1.
Molecular alterations with approved therapies in non-small-cell lung cancer
| Gene/protein alteration | Approved therapy |
|---|---|
| EGFR exon 19 deletion or exon 21 L858R mutations | Afatinib*, dacomitinib*, erlotinib*, gefitinib*, osimertinib* |
| EGFR S678I, L861Q, G719X mutations | Afatinib* |
| EGFR exon 20 insertions | Avimantanab⁎⁎, mobocertinib⁎⁎ |
| BRAF V600E mutation | Dabrafenib*, trametinib* |
| ALK rearrangement | Alectinib*, brigatinib*, ceritinib*, crizotinib*, lorlatinib* |
| ROS1 rearrangement | Crizotinib*, entrectinib* |
| RET rearrangement | Pralsetinib*, selpercatinib* |
| NTRK rearrangements | Entrectinib*, larotrectinib* |
| MET exon 14 skipping mutation | Capmatinib⁎⁎, crizotinib*, tepotinib⁎⁎ |
| KRAS G12C mutation | Sotorasib⁎⁎ |
only FDA-approved
ALK, anaplastic lymphoma kinase; BRAF, B-Raf proto-oncogene; EGFR, epidermal growth factor receptor; HER2, human epidermal growth factor 2; KRAS, kirsten rat sarcoma viral oncogene homologue; MET, mesenchymal-epithelial transition; NRG1, neuregulin-1; NTRK, neurotrophic tyrosine receptor kinase 1; RET, rearranged during transfection; ROS1, C-ros oncogene 1
Given the established efficacy of targeted therapies in tumors harboring oncogenic drivers, the European and American guidelines recommend molecular testing for all advanced NSCLCs of non-squamous histology, particularly those with probable or definite adenocarcinoma, as in patients with non-adenocarcinoma histology, with low tobacco exposure, young age, or small specimen biopsy regardless of the performance status, to retrieve the most complete information to define the first-line treatment.4–9
Despite the recognition of the relevance of molecular characterization in the management of NSCLC, several challenges still need to be addressed and overcome in the clinical practice to ensure a complete and fast molecular assessment. An expert panel of pulmonologists and oncologists dedicated to thoracic Oncology convened to debate the molecular characterization of NSCLC at diagnosis and progression and identify key aspects to improve patient outcomes, highlighting the current evidence on NSCLC molecular profiling and discussing its benefits and challenges.
The following aspects were identified as most relevant for standardizing and optimizing the molecular diagnostic process considering both the laboratory and clinical approach:
- •
Reflex testing has the advantage of optimizing sample management. It reduces the time until treatment initiation and should be performed by the pathologist after histological assessment.5
- •
Molecular analysis should include a comprehensive gene panel, ideally a targeted multiplex next-generation sequencing (NGS) panel including point mutations, deletions, and rearrangements. Given the increasing number of mutations with potential clinical impact, NGS allows to optimize sample processing and simultaneously screen for several genes, with high throughput and sensitivity and low cost per test.10–13 Although NGS is a costly technique for most centres, it will predictably become more accessible in the future, as demonstrated in studies exploring the cost-effectiveness of the method.10
- •
Biomarkers assessed should target genomic drivers with approved therapies, including the epidermal growth factor receptor (EGFR), the anaplastic lymphoma kinase (ALK), the C-ros oncogene 1 (ROS1), the rearranged during transfection (RET), and the B-Raf proto-oncogene (BRAF). In addition, given the fast pace of therapeutic progress, molecular alterations with targeted therapies in advanced stages of development or likely to become therapeutic targets in the short term should also be assessed, specifically those in the human epidermal growth factor 2 (HER2), neurotrophic tyrosine receptor kinase (NTRK), mesenchymal-epithelial transition (MET), and Kirsten rat sarcoma viral oncogene homologue (KRAS). This approach is in accordance with the European Society for Medical Oncology (ESMO) recommendations for the use of NGS in the clinical practice,10 and allows the treatment in routine clinical practice as well as access to ongoing clinical trials and early access programs.5,6
- •
In specific cases of very symptomatic patients with aggressive disease and urgent need for treatment, rapid tests can be considered to define the first-line treatment, namely polymerase chain reaction (PCR) to detect EGFR mutations and immunohistochemistry or FISH to detect ALK and ROS1 rearrangements, while maintaining NGS ongoing.
- •
In patients without sufficient tumor tissue to undergo molecular testing who are ineligible for rebiopsy, liquid biopsy can be considered to identify therapeutic targets. Liquid biopsy has several advantages, like avoiding the potential complications of tissue biopsy and allowing serial monitoring. In addition, it can provide a complete and real-time molecular profile, with information about clonal evolution and dynamic modifications within the tumor.14 The clinical use of liquid biopsy in detecting EGFR mutations in plasma from advanced NSCLC patients has been validated6,13,15–21 and is currently being assessed for other oncogenic drivers, as ALK, BRAF, ROS1, MEK, and HER2.13,22–25
- •
Despite the significant improvements in patient outcomes achieved with EGFR-tyrosine kinase inhibitors (TKI), most patients acquire resistance and develop progressive disease within 10–12 months of treatment, limiting its long-term efficacy. This is particularly true when considering first- and second-generation TKIs.26 The most commonly acquired resistance mutation to first- and second-generation EGFR-TKIs is the T790M mutation in EGFR exon 20, identified in around 50–60% of cases.27–29 Other acquired resistance mechanisms to these inhibitors include alternative pathway activation through c-Met amplification, HER2 activation, and PIK3CA and BRAF mutations and histological transformation.30 T790M confers resistance to gefitinib, erlotinib, and afatinib, and its detection allows the use of the third-generation EGFR-TKI osimertinib in second line.24 At disease progression, rebiopsy should be considered to look for targetable resistance mechanisms. In the setting of EGFR-mutated disease, liquid biopsy can be the first step, as it is more accessible and less invasive than other methods. In cases of progression to third-generation TKIs, NGS is preferred to single-detection testing.8 Tissue biopsy should be considered in cases of negative or inconclusive liquid biopsy, progression to third-generation TKIs, and rapidly progressive disease, to investigate histological transformation.8
- •
The average time between histological diagnosis and getting the molecular test result is heterogeneous among institutions, but ideally should not exceed two weeks.6 Minimizing bureaucratic issues and adopting reflex testing can reduce this time. Irrespective of the test being performed in-house or in an external laboratory, timely retrieval of results should be ensured.
- •
The molecular study report should be presented in a simplified and systematic way and include the molecular test results (gene panel used and genomic changes identified and respective allele frequencies), their clinical interpretation given the available evidence, therapeutic options, and ongoing clinical trials (which should be regularly updated).31
- •
Molecular study results should always be discussed within a multidisciplinary context, to allow complete and thorough data analysis. Defining a molecular tumor board is a recommended best practice that all centers should adopt.8
The management of NSCLC remains challenging, and the integration of data from predictive biomarkers in routine clinical practice can contribute to an optimal, individualized patient approach, particularly given the rapid emergence of effective targeted therapies. When considering molecular biomarker testing, the choice of the biomarker panel, target population, testing approach, and turnaround time are key issues that, when properly addressed, can improve the survival outcomes of NSCLC patients.
Medical writing assistance, supported financially by Boehringer Ingelheim Portugal, was provided by Prime Focus, during the preparation of this article