To investigate the differences in mRNA and protein expressions of MDM2 (mouse double minute 2 homolog) and P73 in cancer and cancer-adjacent tissues in patients with non-small-cell lung carcinoma (NSCLC).
Materials and methodsWe compared the protein expressions of MDM2 and P73 in lung cancer and cancer-adjacent tissues in NSCLC patients by IHC (immunohistochemistry) and WB (Western blot). We divided the NSCLC patients into two subgroups, adenocarcinoma and squamous carcinoma. The mRNA expressions of two main isoforms of P73, TAP73 and DNP73, as well as the ratio of DNP73/TAP73 were analyzed by qPCR (quantitative real-time PCR) in the two tissues in all NSCLC patients and in patients with adenocarcinoma or squamous carcinoma, respectively.
ResultsWB results did not show significant differences in MDM2 and P73 protein expressions in lung cancer and cancer-adjacent tissues. However, IHC results indicated that MDM2 expression significantly increased in cancer tissues in female patients, but not male patients. In addition, TAP73 mRNA expression significantly increased in cancer tissues in all NSCLC patients (p=0.002) and in patients with adenocarcinoma (p=0.01); while there was no significant difference in DNP73 mRNA expression. Hence the fold-change of DNP73/TAP73 ratio significantly decreased (p=0.0003) in cancer tissues in all NSCLC patients and in patients with adenocarcinoma.
ConclusionsTAP73 mRNA expression significantly increased in cancer tissues than cancer-adjacent tissues in all NSCLC patients and in patients with adenocarcinoma. Meanwhile, the fold-change of DNP73/TAP73 ratio was similar to TAP73. MDM2 protein expression significantly increased in cancer tissues in female NSCLC patients.
Lung cancer is a malignant tumor with the highest morbidity and mortality, and is a serious threat to human health. The etiology of lung cancer is the interaction of environment factors (smoking,1,2 air pollution, ionizing radiation,3 and diet4) and genetic factors.5 Lung cancer can be classified into two major types, SCLC (small cell lung cancer) and NSCLC (non-small cell lung cancer), according to the histopathology. The most common types of NSCLC are SC (squamous carcinoma), adenocarcinoma, and large cell carcinoma.6 Adenocarcinoma accounts for 50% of all lung cancer cases. SC is more common in elderly men and is correlated with smoking. SC is sensitive to CT (chemotherapy) and RT (radiotherapy) treatments. The best treatment for patients with SC is surgical approach in a combination of CT and RT7 and the five-year survival rate is relative high in this context.8 Adenocarcinoma is more frequently observed in female patients and is not always with smoking. The morbidity of adenocarcinoma has risen in recent years and it has become the main type of lung cancer in some countries. Although the therapeutic methods have been improved, the overall-survival rate of lung cancer has not improved in recent years.9 Hence, a deeper understanding of the etiology of lung cancer is necessary for the development of new therapeutic approaches and the treatment of lung cancer.
TP53 is a classical tumor-suppressor gene10 and is frequently altered in majority of the human cancers,11 resulting in the expression of mutant P53 proteins with single-amino-acid substitutions within the DNA-binding domain (DBD).12 Therefore, TP53 plays an important role in maintaining the genome integrity.13P73 and P63 are two homologs of TP53. Unlike TP53, P63 and P73 regulate developmental processes rather than participate in the control of genome stability.14P73 is located on human chromosome 1p36.3, and is consisted of 13 exons and 12 introns. It has been reported that P73 plays an important role in cancers.15 P73 is involved in the control of programmed cell death,16 and can be used as an indicator of cancer prognosis.17P73 mutation is often resulted in a variety of tumors, including neurocytoma, CRC (colorectal cancer) and breast cancer.18,19
P73 encodes two isoforms, TAP73 (transcriptionally active P73) and DNP73 (dominant negative P73).20 Studies show that P73 mRNA expression is higher in cancer tissues than in healthy tissues, suggesting that P73 might be a oncogene.21 Evidence indicates that TAP73 can suppress tumors formation while DNP73 can promote tumor formation.22 Studies have found that TAP73 and DNP73 are overexpressed in ovarian cancer, hepatocellular carcinoma and colon cancer, and their expression levels are correlated with the development and prognosis of cancers.22–25 Accumulating evidence suggests that the overexpression of DNP73 transcript is associated with adverse prognosis and chemotherapy failure in several human tumors.26 High DNP73/TAP73 ratio is associated with poor prognosis in acute promyelocytic leukemia (APL).27 The expression of TAP73 and DNP73 can be elevated simultaneously in lung cancer. Hence, TAP73 and DNP73 interact with each other and play complex roles in regulating the proliferation and apoptosis of lung cancer.28
MDM2 is located on human chromosome 12q14.3-q15, and is one of the principal ubiquitin ligases that are responsible for P53 degradation.29,30 MDM2 can regulate the activity, stability and function of P5331 and can also interact with P73.32,33 In MDM2-P53 system, P53 activation induces MDM2 transcription; while MDM2 activation inhibits P53 activity by binding to its activated area of transcription.34 However, it is unclear whether MDM2 can regulate P73 activity.
Studies show that MDM2 and P73 can form heterodimers in vivo or in vitro. MDM2 does not promote P73 degradation,35 but it can suppress P73 protein expression by binding to the N terminal of the p300/CBP; while P73 can stimulate the expression of endogenous MDM2. Hence, MDM2 is a negative feedback regulator of P73, and form a negative feedback loop with P73.14 MDM2-P73 system plays an important role in the development of lung cancer.36 It has been reported that MDM2 overexpression and P73 deficiency can induce genome instability and tumor development.37,38
To date, no study has reported the expressions of MDM2 and P73 in different types of lung cancers. Hence, in this study, we investigated the relationship between MDM2 and P73 in lung cancers, as well as the functions of TAP73 and DNP73 in the development and prognosis of lung cancer.
Materials and methodsPatients and materialsWe calculated the estimated sample size based on our preliminary data. We selected 45 patients with lung cancer in our hospital from June 2016 to October 2016. The inclusion criteria included: (1) The patients had not received chemotherapy (CT), radiotherapy (RT), biological drug treatment (drugs that could bind to the specific cancer site and kill the cancer cells) and surgery; (2) the patients did not have other tumors (such as carcinoma); (3) the patients were suitable for surgery; (4) the patients did not have other non-cancer diseases according to http://geneontology.org/ (such as aquaphobia). We collected the samples of cancer tissues and cancer-adjacent tissues from all 45 patients. The lung cancer tissues were further divided into different sub-groups according to the histopathology, including 10 cases of squamous carcinoma, 31 cases of adenocarcinoma and 4 cases of other cancer types. Meanwhile, we also collected the basic and important information of all the patients.
This study was approved by the Hospital Ethics Committee (No. 2015034), and all the patients signed the informed consent.
Methods and statistical analysisMethodsWe measured the MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues by IHC (immunohistochemistry) and WB (Western blot), respectively. The main reagents and instruments were shown in Appendix Table 1 (Table A.1).
We measured the MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues by IHC (immunohistochemistry) and WB (Western blot), respectively. The main reagents and instruments were shown in Appendix Table 1 (Table A.1).
IHCThe frozen tissues were dehydrated at room temperature and fixed with 4% paraformaldehyde for 15min. Tissues were paraffin-embedded and sectioned. The sections were incubated in 5% H2O2 at room temperature for 15min. The sections were incubated in EDTA for 3min at 140°C, washed 3 times in PBS, 5min each. The sections were blocked with 5% goat serum for 20min at 37°C and incubated with the primary antibody (1:100) overnight at 4°C. After washing, the sections were incubated with biotinylated secondary antibody for 30min at room temperature. Sections were washed 4 times in PBS and dehydrated with sequential ethanol gradients (75%, 80%, and 100%). Images were acquired by optical microscopy.
IPP (Image-pro plus 6.0) software was used to analyze the IHC images. The ratio of region of interest to overall area was calculated to analyze the difference between MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues. The main principle and process were as follows: images from five different fields in each tissue were randomly acquired. The density of the background was adjusted to distinguish the background and target area. The region of interest (ROI, the brown staining area) on each image was defined and the total area was measured. The values from the five images were exported and the mean values were calculated.
The accumulated IOD (Integrated optical density) of the brown background in a selected field was measured, and the mean IOD was calculated by the formula: Mean IOD=IOD/(Total area of the selected field).
WBThe proteins were extracted from the cancer and cancer-adjacent tissues of patients using RIPA lysis Buffer. The protein concentration was measured using BCA (Bicinchoninic acid) Kit according to the manufacturer's instructions. 32μg sample was loaded onto 10% SDS-PAGE, run at 90V for 20min, and 120V for 50min. After electrophoresis, protein samples were transferred onto PVDF membranes (0.45μm). The membranes were incubated in ponceau and the protein bands were observed. The membranes were blocked in 5% BSA-TBST for 1h, and then incubated with primary antibodies (1:500) overnight at 4°C. Next day, the membranes were washed 3 times with TBST, 10min each. The membranes were incubated with secondary antibodies (1:10,000) for 40min at room temperature. After washing, the membranes were developed using ECL and exposed to X-ray. Films were scanned by scanner and Gel-Pro analyzer was used to analyze the Greyscale for protein quantification. Gel Image system ver.4.00 (Tanon, China) software was used to analyze the WB outcomes. Beta-actin was used as the internal control. The formula for the calculation of mean gray value was as follows: Mean gray value=object value/internal control value. The experiments were repeated three times and the values were calculated and averaged.
RT-PCR (reverse transcription PCR) and qPCR (quantitative real-time PCR)RT-PCR and qPCR was used to detect the mRNA expressions of TAP73 and DNP73 (two isoforms of P73) in cancer tissues and cancer-adjacent tissues from each patient. The primers were: TAP73 (Amplicon size: 111bp) forward: 5′-GCACCACGTTTGAGCACCTCT-3′, reverse: 5′-GCAGATTGAACTGGGCCATGA-3′; DNP73 (Amplicon size: 123bp) forward: 5′ACT AGC GCG GAG CCT CTC CC-3′, reverse: 5′T GC T CA GCA GAT GAA CTG G-3′; H-ACTB (Amplicon size: 127bp) forward: AGCACAATGAAGATCAAGATCAT, reverse: ACTCGTCATACTCCTGCTTGC. Other regents and instruments were shown in Appendix Table 1 (Table A.1).
RT-PCR and qPCR was used to detect the mRNA expressions of TAP73 and DNP73 (two isoforms of P73) in cancer tissues and cancer-adjacent tissues from each patient. The primers were: TAP73 (Amplicon size: 111bp) forward: 5′-GCACCACGTTTGAGCACCTCT-3′, reverse: 5′-GCAGATTGAACTGGGCCATGA-3′; DNP73 (Amplicon size: 123bp) forward: 5′ACT AGC GCG GAG CCT CTC CC-3′, reverse: 5′T GC T CA GCA GAT GAA CTG G-3′; H-ACTB (Amplicon size: 127bp) forward: AGCACAATGAAGATCAAGATCAT, reverse: ACTCGTCATACTCCTGCTTGC. Other regents and instruments were shown in Appendix Table 1 (Table A.1).
I. RNA isolation50mg tissues were pulverized in liquid nitrogen, and transferred into centrifugal tubes. The tissues were homogenized in 1mL Trizol and incubated at room temperature for 5min. 0.2mL trichloromethane was added, vortexed for 10s, and incubated at room temperature for 5min. The samples were centrifuged at 12,000rpm for 15min at 4°C, and 550μL supernatants were collected. After adding the same volume of isopropanol, the samples were incubated at −20°C for 20min, and then centrifuged at 12,000rpm for 15min at 4°C. The RNA pellet was washed with 1mL 75% ethyl alcohol and centrifuged at 12,000rpm for 15min at 4°C. The supernatant was removed and the RNA samples were air dry for 5min. RNA was dissolved in 30μL RNase-free water.
II. RT-PCRAfter Dnase treatment RNA was reverse-transcribed into cDNA using HiFiScript kits. 10-μL reaction system included 1μg RNA Template, 0.5μL gDNA Eraser and 1μL 10×g DNA Eraser Buffer. Samples were heated at 42°C for 2min and cooled on ice. 1μL HiFiScript (200U/μl), 1μL Primer Mix, 4μL 5× RT Buffer and 4μL RNase-free ddH2O were added. The thermal cycles were 42°C for 50min, 85°C for 5min, and 4°C forever.
III. Real-time PCRReal-time PCR was carried out with KAPA SYBR FAST qPCR Kit Master Mix (2×) (KAPA Biosystems, KK4601). The 10μL reaction system included 5μL PCR Master Mix (2×), 0.2μL mRNA forward primers (10μM), 0.2μL mRNA reverse primers (10μM), 1μL cDNA, 0.2μL Dye (50×), and 3.4μL ddH2O.
The reaction protocol was 3min at 95°C activation, 40 cycles of 3s at 95°C and 20s at 60°C. Melting curve was constructed in the range of 60–95°C. The original data, amplification curve and solubility curve were exported to quantification software. The relative expression levels of target genes were analyzed using 2−ΔΔCt method. The fold-change of mRNA expression in cancer tissues relative to cancer-adjacent tissues was compared.
Statistical analysisSPSS 19.0 software was used to analyze all the data. Mean value and standard error were used to present MDM2 and P73 protein expressions, as well as TAP73 and DNP73 mRNA expressions. One-way ANOVA (one-way analysis of variance) with Bonferroni–Dunnett corrections were used for multiple-group comparisons. p<0.05 indicated a statistical significance.
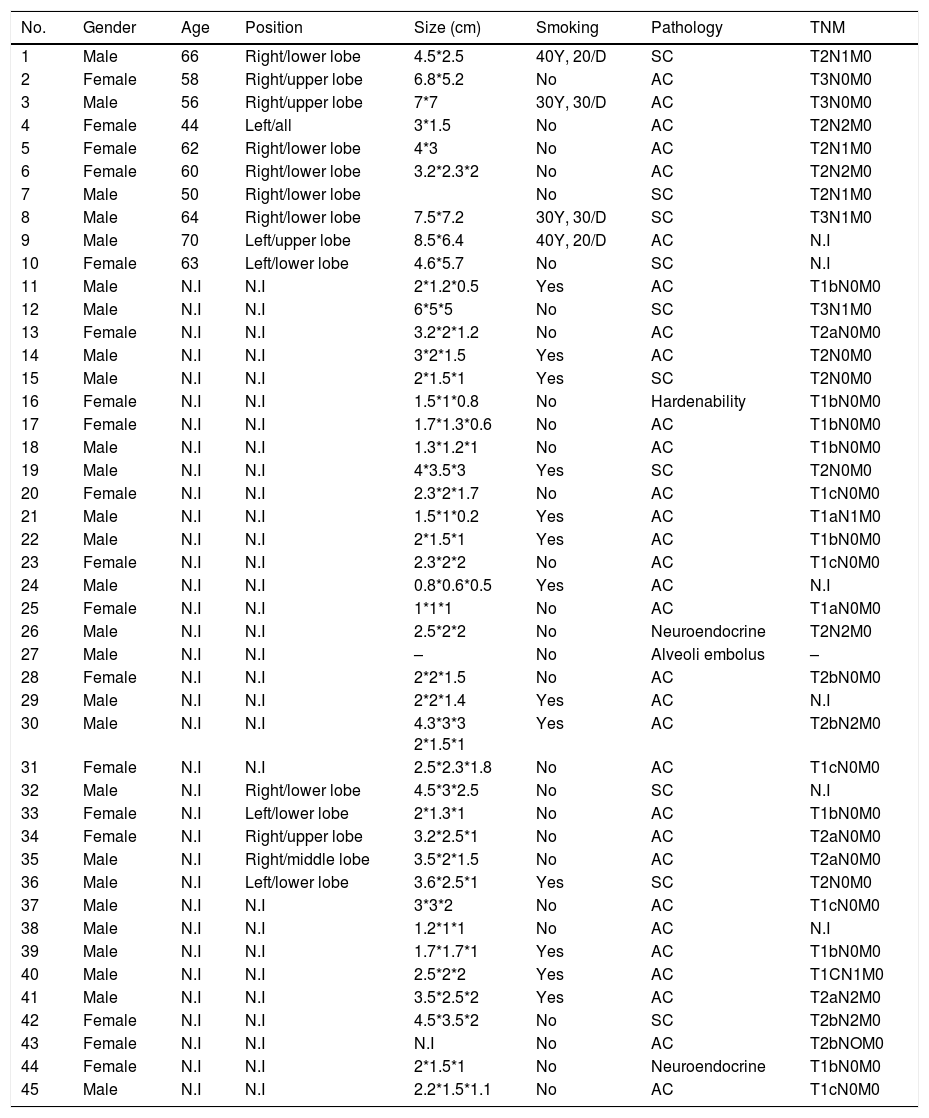
ResultsThe summary of the patients’ information was as follows: (1) the age of the patients ranged from 40 to 70 years old. (2) 27 were male and 18 were female. (3) The cancer types were roughly divided into adenocarcinoma (22.2%), squamous carcinoma (68.9%) and others (8.9%). (4) TNM tumor stages varied among patients; however, no metastasis (M=0) was found in all the patients we analyzed. The details of the patients’ information were shown in Table 1.
Characteristics of the patients.
| No. | Gender | Age | Position | Size (cm) | Smoking | Pathology | TNM |
|---|---|---|---|---|---|---|---|
| 1 | Male | 66 | Right/lower lobe | 4.5*2.5 | 40Y, 20/D | SC | T2N1M0 |
| 2 | Female | 58 | Right/upper lobe | 6.8*5.2 | No | AC | T3N0M0 |
| 3 | Male | 56 | Right/upper lobe | 7*7 | 30Y, 30/D | AC | T3N0M0 |
| 4 | Female | 44 | Left/all | 3*1.5 | No | AC | T2N2M0 |
| 5 | Female | 62 | Right/lower lobe | 4*3 | No | AC | T2N1M0 |
| 6 | Female | 60 | Right/lower lobe | 3.2*2.3*2 | No | AC | T2N2M0 |
| 7 | Male | 50 | Right/lower lobe | No | SC | T2N1M0 | |
| 8 | Male | 64 | Right/lower lobe | 7.5*7.2 | 30Y, 30/D | SC | T3N1M0 |
| 9 | Male | 70 | Left/upper lobe | 8.5*6.4 | 40Y, 20/D | AC | N.I |
| 10 | Female | 63 | Left/lower lobe | 4.6*5.7 | No | SC | N.I |
| 11 | Male | N.I | N.I | 2*1.2*0.5 | Yes | AC | T1bN0M0 |
| 12 | Male | N.I | N.I | 6*5*5 | No | SC | T3N1M0 |
| 13 | Female | N.I | N.I | 3.2*2*1.2 | No | AC | T2aN0M0 |
| 14 | Male | N.I | N.I | 3*2*1.5 | Yes | AC | T2N0M0 |
| 15 | Male | N.I | N.I | 2*1.5*1 | Yes | SC | T2N0M0 |
| 16 | Female | N.I | N.I | 1.5*1*0.8 | No | Hardenability | T1bN0M0 |
| 17 | Female | N.I | N.I | 1.7*1.3*0.6 | No | AC | T1bN0M0 |
| 18 | Male | N.I | N.I | 1.3*1.2*1 | No | AC | T1bN0M0 |
| 19 | Male | N.I | N.I | 4*3.5*3 | Yes | SC | T2N0M0 |
| 20 | Female | N.I | N.I | 2.3*2*1.7 | No | AC | T1cN0M0 |
| 21 | Male | N.I | N.I | 1.5*1*0.2 | Yes | AC | T1aN1M0 |
| 22 | Male | N.I | N.I | 2*1.5*1 | Yes | AC | T1bN0M0 |
| 23 | Female | N.I | N.I | 2.3*2*2 | No | AC | T1cN0M0 |
| 24 | Male | N.I | N.I | 0.8*0.6*0.5 | Yes | AC | N.I |
| 25 | Female | N.I | N.I | 1*1*1 | No | AC | T1aN0M0 |
| 26 | Male | N.I | N.I | 2.5*2*2 | No | Neuroendocrine | T2N2M0 |
| 27 | Male | N.I | N.I | – | No | Alveoli embolus | – |
| 28 | Female | N.I | N.I | 2*2*1.5 | No | AC | T2bN0M0 |
| 29 | Male | N.I | N.I | 2*2*1.4 | Yes | AC | N.I |
| 30 | Male | N.I | N.I | 4.3*3*3 2*1.5*1 | Yes | AC | T2bN2M0 |
| 31 | Female | N.I | N.I | 2.5*2.3*1.8 | No | AC | T1cN0M0 |
| 32 | Male | N.I | Right/lower lobe | 4.5*3*2.5 | No | SC | N.I |
| 33 | Female | N.I | Left/lower lobe | 2*1.3*1 | No | AC | T1bN0M0 |
| 34 | Female | N.I | Right/upper lobe | 3.2*2.5*1 | No | AC | T2aN0M0 |
| 35 | Male | N.I | Right/middle lobe | 3.5*2*1.5 | No | AC | T2aN0M0 |
| 36 | Male | N.I | Left/lower lobe | 3.6*2.5*1 | Yes | SC | T2N0M0 |
| 37 | Male | N.I | N.I | 3*3*2 | No | AC | T1cN0M0 |
| 38 | Male | N.I | N.I | 1.2*1*1 | No | AC | N.I |
| 39 | Male | N.I | N.I | 1.7*1.7*1 | Yes | AC | T1bN0M0 |
| 40 | Male | N.I | N.I | 2.5*2*2 | Yes | AC | T1CN1M0 |
| 41 | Male | N.I | N.I | 3.5*2.5*2 | Yes | AC | T2aN2M0 |
| 42 | Female | N.I | N.I | 4.5*3.5*2 | No | SC | T2bN2M0 |
| 43 | Female | N.I | N.I | N.I | No | AC | T2bNOM0 |
| 44 | Female | N.I | N.I | 2*1.5*1 | No | Neuroendocrine | T1bN0M0 |
| 45 | Male | N.I | N.I | 2.2*1.5*1.1 | No | AC | T1cN0M0 |
No., number; TNM, topography, lymph node and metastasis; Y, years; D, day; SC, squamous carcinoma; AC, adenocarcinoma; N.I: no information.
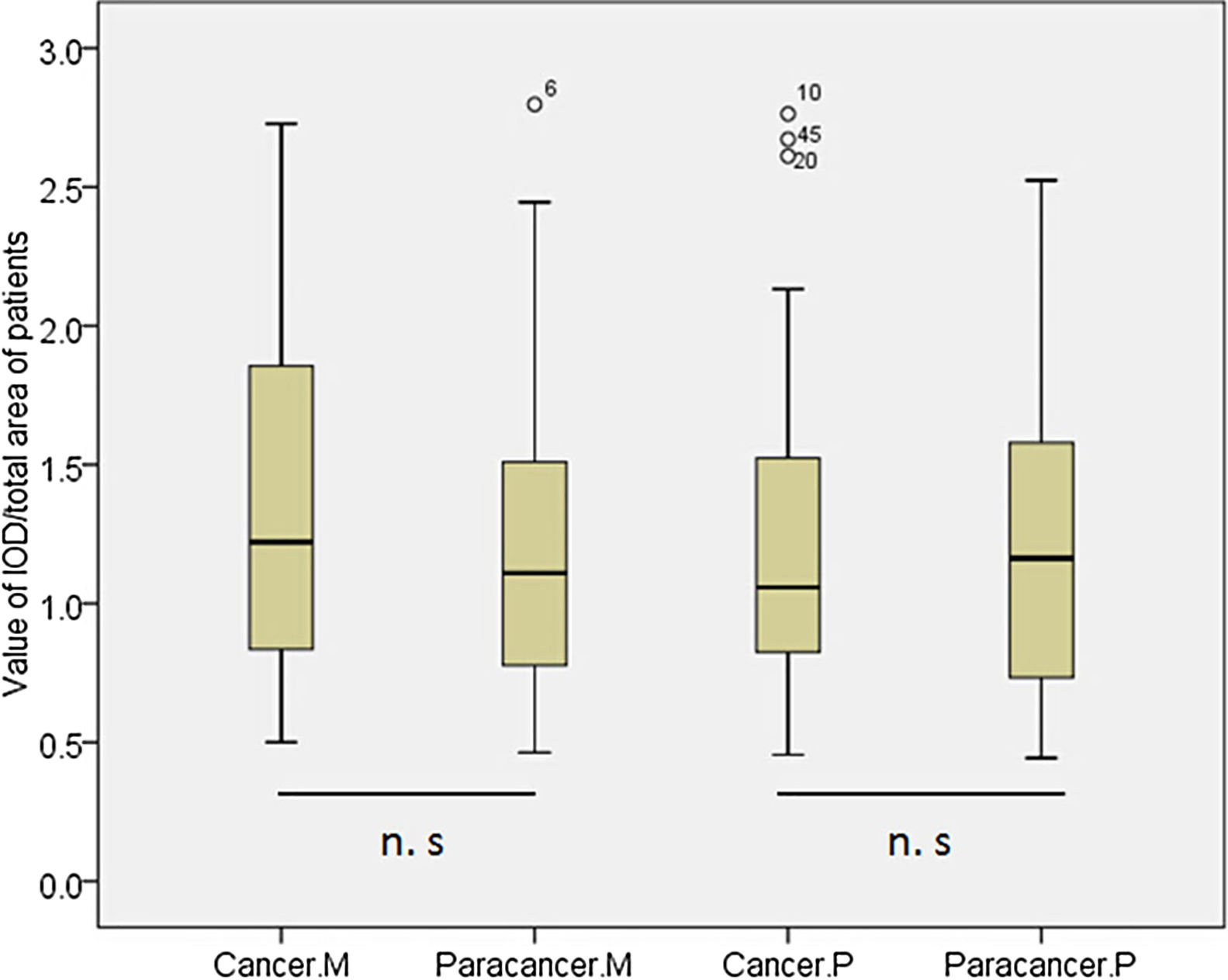
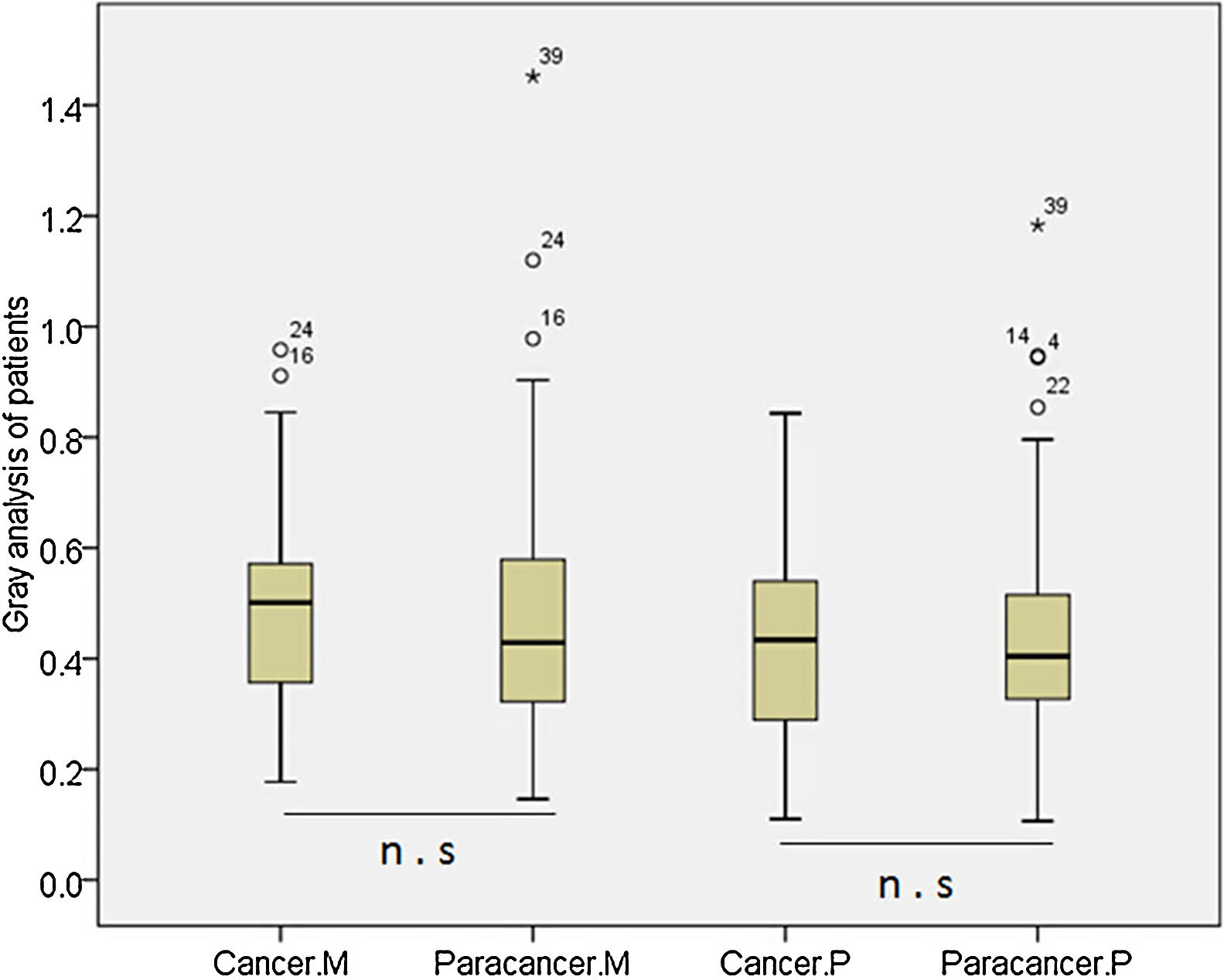
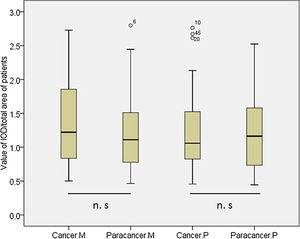
According to the quantitative analysis using IPP software, we compared MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues. We found that MDM2 (p>0.05) and P73 (p>0.05) expressions were similar in cancer and cancer-adjacent tissues in all patients (Appendix Fig. A.1). We also compared the MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues in patients with squamous carcinoma or adenocarcinoma, respectively. However, we did not find any significant difference.
According to the quantitative analysis using IPP software, we compared MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues. We found that MDM2 (p>0.05) and P73 (p>0.05) expressions were similar in cancer and cancer-adjacent tissues in all patients (Appendix Fig. A.1). We also compared the MDM2 and P73 protein expressions in cancer and cancer-adjacent tissues in patients with squamous carcinoma or adenocarcinoma, respectively. However, we did not find any significant difference.
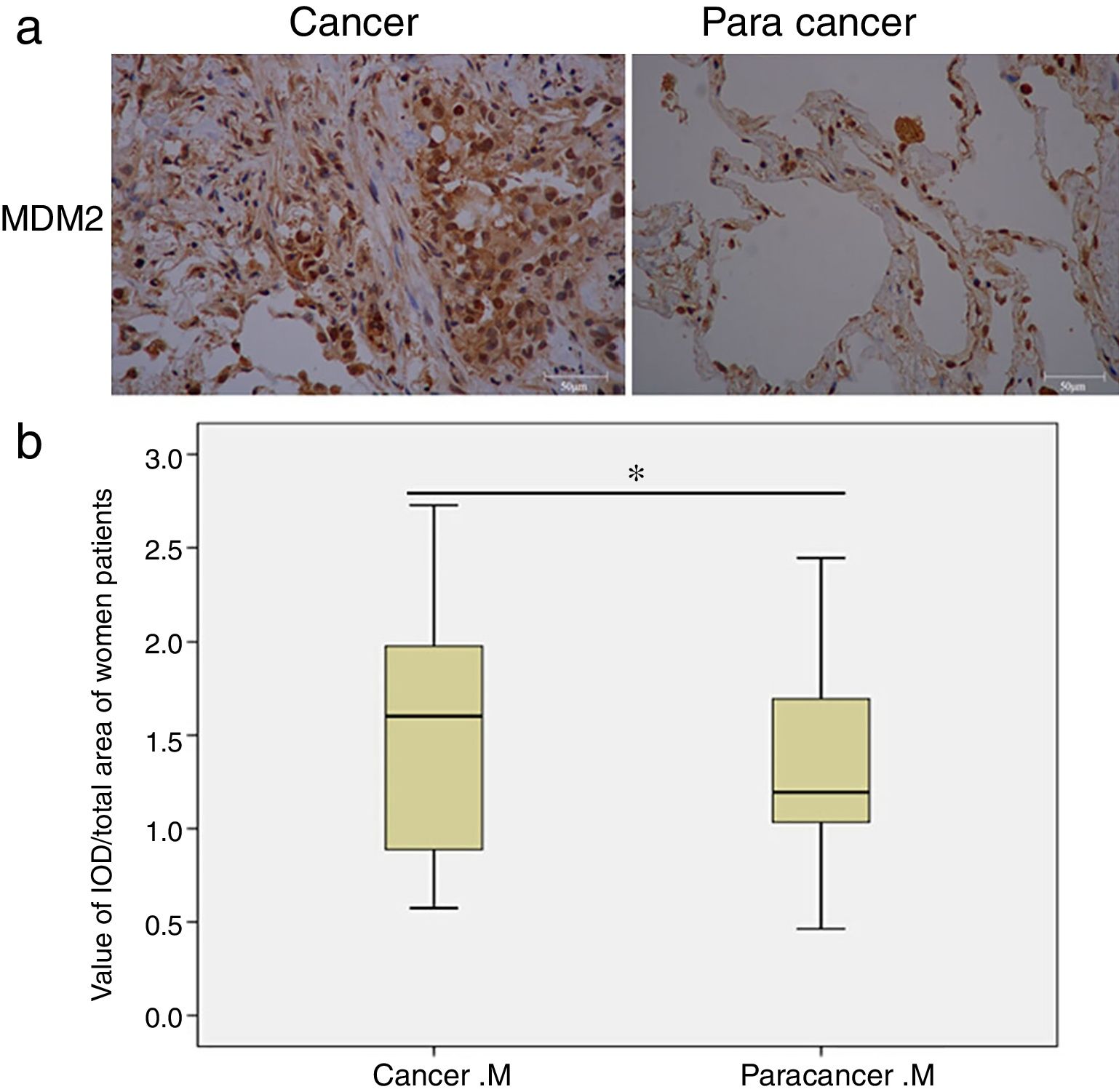
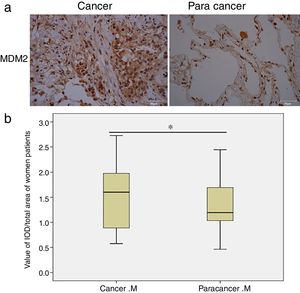
Next, we analyzed the MDM2 and P73 protein expressions based on gender and smoking history. Interestingly, we found that MDM2 expression significantly increased in cancer tissues only in the female patients (p=0.01, Fig. 1), but not in the male patients. Moreover, we found that the MDM2 was mainly expressed in the nucleus in the cancer-adjacent tissues; while MDM2 was simultaneously expressed in nucleus and cytoplasm in the cancer tissues. There was no significant difference in P73 expression in male and female patients. Moreover, there was no statistical significance in MDM2 and P73 expressions in smoking and non-smoking groups.
MDM2 expression in female NSCLC patients detected by IHC. (a) Representative images of MDM2 expression in cancer and cancer-adjacent tissues from a female NSCLC patient; (b) box-plots show the quantitative analysis of MDM2 expression in cancer and cancer-adjacent tissues from all female NSCLC patients. The results indicate that MDM2 expression is significantly increased in cancer tissues of women patients. M: MDM2. *p<0.05, indicating a significant difference.
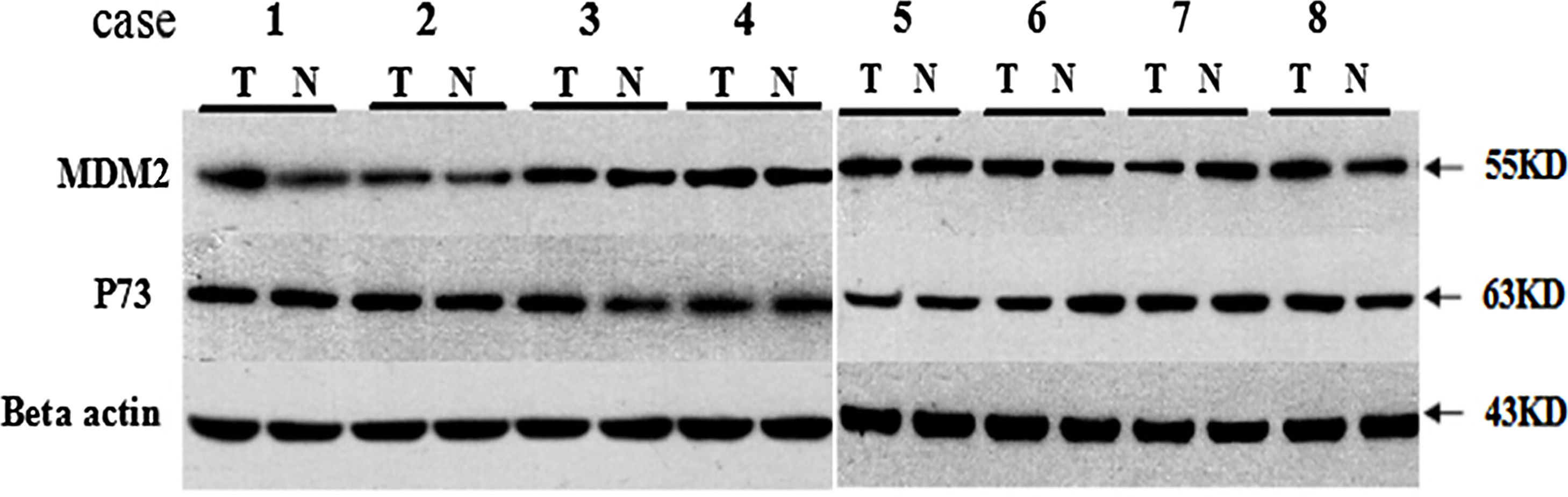
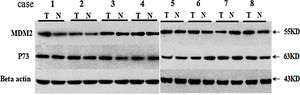
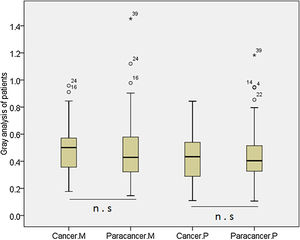
We analyzed the MDM2 and P73 protein expressions using WB and the representative WB images are shown in Appendix Fig. A.2. We found that there was no significant difference in the MDM2 and P73 protein expressions between cancer and cancer-adjacent tissues in all patients (p>0.05, Appendix Fig. A.3). We also analyzed the MDM2 and P73 protein expressions in patients with adenocarcinoma or squamous carcinoma, respectively. However, we did not find any significant diffidence.
We analyzed the MDM2 and P73 protein expressions using WB and the representative WB images are shown in Appendix Fig. A.2. We found that there was no significant difference in the MDM2 and P73 protein expressions between cancer and cancer-adjacent tissues in all patients (p>0.05, Appendix Fig. A.3). We also analyzed the MDM2 and P73 protein expressions in patients with adenocarcinoma or squamous carcinoma, respectively. However, we did not find any significant diffidence.
Next, we analyzed the MDM2 and P73 protein expressions based on gender and smoking history. However, there was no significant difference in MDM2 and P73 expressions in male and female patients, and smoking and non-smoking groups.
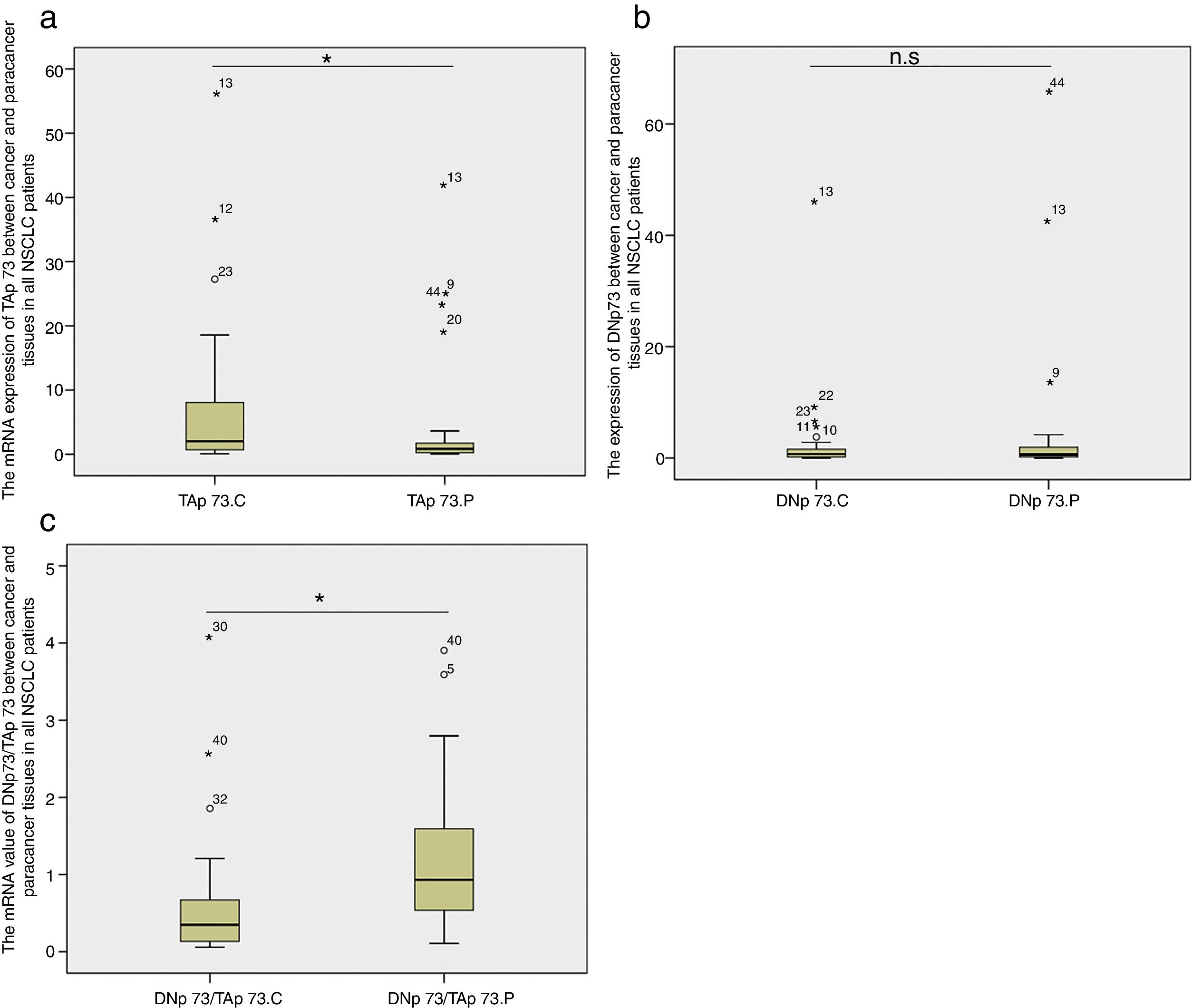
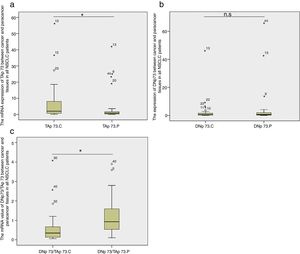
TAP73 and DNP73 mRNA expressionsWe compared the fold-change of TAP73 and DNP73 mRNA expression in cancer tissues relative to cancer-adjacent tissues in all lung cancer patients. We found that there was a significant increase in TAP73 mRNA expression in cancer tissues (p=0.035, Fig. 2a); while there was no significant difference in DNP73 mRNA expression (p=0.415, Fig. 2b). The ratio of DNP73/TAP73 significantly decreased in cancer tissues (p=0.0003) (Fig. 2c).
Box-plots show the mRNA expression of TAP73 (a), DNP73 (b) and the ratio of DNP73/TAP73 (c) between cancer and cancer-adjacent tissues in all NSCLC patients. n. s: no significant; C; cancer; P: paracancer; small circles: abnormal values; small starlets: significantly abnormal values; figures: the number of abnormal values.
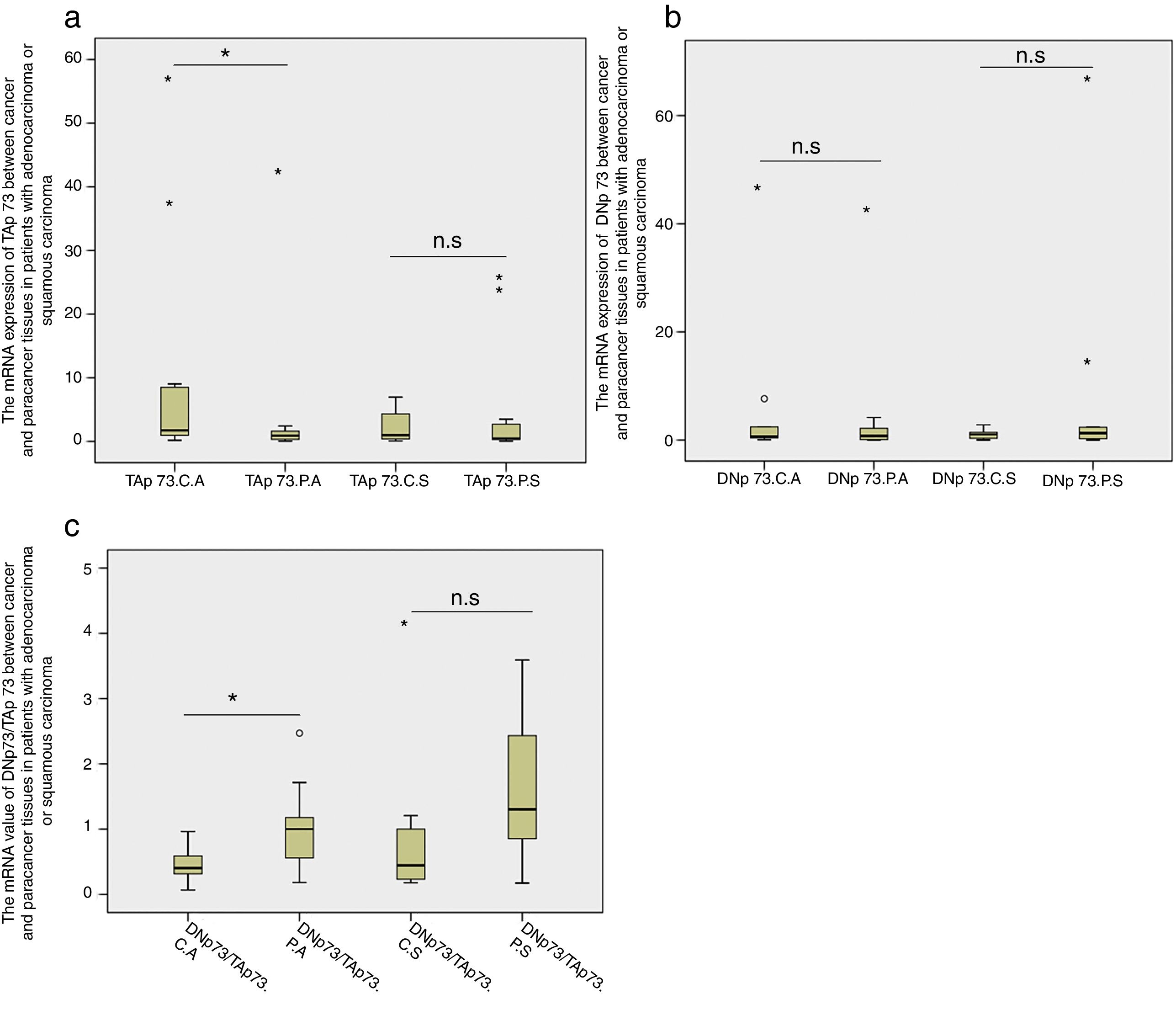
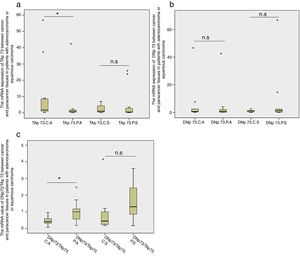
Next we compared the TAP73 and DNP73 mRNA expressions in patients with adenocarcinoma or squamous carcinoma, respectively. We found that TAP73 mRNA expression was significantly higher in the cancer tissues in patients with adenocarcinoma (p=0.01, Fig. 3a) while the TAP73 mRNA expression was similar between cancer and caner-adjacent tissues in patients with squamous carcinoma. Moreover, there was no significant difference in the DNP73 mRNA expression in patients with adenocarcinoma or squamous carcinoma (Fig. 3b). The fold changes of the ratio of DNP73/TAP73 significantly decreased in cancer tissues in patients with adenocarcinoma (p=0.002), but not in patients with squamous carcinoma (Fig. 3c).
DiscussionIn this study we compared the differences in MDM2 and P73 expressions between the cancer and cancer-adjacent tissues in patients with NSCLC. Interestingly, TAP73 mRNA expression, an isoform of P73, significantly increased in the cancer tissues in all NSCLC patients and in patients with adenocarcinoma; while there was no difference in DNP73 mRNA expression. Therefore, fold changes of the ratio of DNP73/TAP73 significantly decreased in cancer tissues in all NSCLC patients and in patients with adenocarcinoma.
Some studies show that TAP73 is a tumor-suppressor gene.35 Irwin MS et al.39 demonstrated that TAP73 can trans-activate P53 target genes, such as Bax, Puma, and P21, inducing apoptosis and cell cycle arrest; however, Deepa Subramanian reported that TAP73 plays a vital role in activation of activator protein-1 (AP-1) target genes, leading to enhanced activation of other AP-1family members and increased cellular growth.40 These results suggest that TAP73 may have different functions in different cells. Moreover, we found that TAP73 mRNA expression significantly increased in cancer tissues in patients with adenocarcinoma, but not in patients with squamous carcinoma. These results may reflect the heterogenic pathology of different types of tumors. Squamous carcinoma is the most common type of NSCLC, and is highly associated with smoking, which is different from adenocarcinoma.
We speculate that elevated TAP73 mRNA expression in cancer tissues may change the interaction between CDK (cell cyclin-dependent kinase) and Cyclins, thereby promoting cell proliferation. However, the underlying mechanisms remain unknown. Moreover, as isoforms of P73, TAP73 and DNP73 exhibit a complex relationship, which have important effects on the function of genes (e.g. P21) that modulate tumor development.
We further compared the MDM2 and P73 protein expressions in the cancer tissues and cancer-adjacent tissues in all NSCLC patients, in patients with adenocarcinoma or squamous carcinoma alone, in male and female patients and in patients with and without smoking history. We found that there was no difference in MDM2 expression when it was analyzed in all NSCLC patients, in patients with adenocarcinoma or squamous carcinoma alone, and in patients with and without smoking history. Interestingly, we found that the MDM2 expression significantly increased in the cancer tissues in only female patients, but not in male patients by IHC, suggesting the expression of MDM2 may be affected by gender. Studies show that women have a higher risk of adenocarcinoma than squamous carcinoma, suggesting that the expression of MDM2 is higher in adenocarcinoma. Moreover, MDM2 overexpression is associated with gynecological cancers,41,42 indicating that the MDM2 expression in tumor tissues might be regulated by estrogen. However, our WB results did not show a significant difference in the MDM2 expression. The discrepancy between the IHC and WB results might be due to the different expression patterns of MDM2 in the cancer tissues and cancer-adjacent tissues. For examples, MDM2 is mainly expressed in the nucleus in the cancer-adjacent tissues, while MDM2 is simultaneously expressed in nucleus and cytoplasm in the cancer tissues. The difference in the MDM2 expression pattern can be detected using IHC, but not by WB. Moreover, we used the whole tissues for WB, some of the areas may not have MDM2 expression, which may cause false negative results. There was no difference in P73 protein expression when it was analyzed in all NSCLC patients or in patients with adenocarcinoma or squamous carcinoma alone, in male and female patients, and in patients with and without smoking history.
Studies show that MDM2 expression is higher in cancer tissues than cancer-adjacent tissues,43 and P73 can act as a tumor-suppressor gene or an oncogene. In lung cancer, Di Vinci A et al.44 found that both DNP73 and TAP73 increased, and the overexpression of TAP73 deteriorates the tumor prognosis, which is similar to the finding of Wen Hong Toh et al. in gastrointestinal carcinomas.45 These findings suggest that a complex regulatory mechanism of P73 may also exist in lung cancer. In our study we demonstrate that TAP73 mRNA expression significantly increased in the lung cancer tissues, which provide new information on the roles of P73 in lung cancer.
It has been reported that the positive rate of MDM2 protein expression is closely correlated with lymph node metastasis, TNM stages, degree of tumor cell differentiation, and tumor recurrence.46 Higashiyama et al.47 demonstrate that MDM2 protein expression detected by IHC can be used as a marker for NSCLC. In our study we found that MDM2 protein expression significantly increased in female lung cancer patients only, which is different from some other studies.43 However, IHC can reveal more details about the expression of MDM2 in cancer and cancer-adjacent tissues than WB. Previous studies have shown that P73 can act with MDM2,35 and MDM2 is involved the occurrence of a variety of tumors.48 However, we did not discover significant correlation between MDM2 and P73 expressions in our study.
This study has some limitations. Firstly, our samples were mainly from patients with adenocarcinoma or squamous carcinoma, and did not include many other types of lung cancer. Therefore, we did not analyze the MDM2 and P73 expressions in other types of lung cancers. Secondly, because we did not find significant difference in P73 protein expression using WB, we did not measure the TAP73 and DNP73 protein expressions by WB. Thirdly, because we did not have enough samples with similar TNM stages, we could not analyze the correlation of MDM2 and P73 with TNM stages. We are continuing to collect samples and will address these limitations in our future studies.
ConclusionIn conclusion, MDM2 protein expression significantly increased in cancer tissues only in female NSCLC patients when it was analyzed by IHC, but not WB. TAP73 mRNA expression significantly increased in cancer tissues in all NSCLC patients and in patients with adenocarcinoma; while there was no change in the DNP73 mRNA expression. Therefore, the fold change of DNP73/TAP73 ratio significantly decreased in cancer tissues in all NSCLC patients and in patients with adenocarcinoma. There was no significant difference in P73 expression between cancer and cancer adjacent tissues. There was no correlation between smoking history and MDM2 and P73 expressions.
Authors’ contributionBW, XL, HL, JG, and TZ participated in data extraction and drafted the manuscript. NZ, YM, HY, and KF carried out the data analysis. LC, ZR, and XT participated in the design of the study. BW, XL, HL, and XT conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All the authors contributed to the interpretation of the results and the proof reading of the manuscript.
Conflicts of interestThe authors have no conflicts of interest to declare.
This research was supported by Beijing Municipal Science &Technology Commission No. Z161100000116075.