Chronic obstructive pulmonary disease (COPD) is a chronic bronchitis (or) emphysema with a high disability and fatality rate. This study aimed to explore the correlation between the six selected single nucleotide polymorphisms (SNPs) and the risk of COPD in the Chinese population.
MethodsThe Agena MassARRAY platform was used to select six SNPs from 629 subjects for genotyping. The correlation between SNPs and COPD risk was evaluated using calculated odds ratios (ORs) and 95% confidence intervals (CIs). Multi-factor dimensionality reduction (MDR) was performed to analyze the impact of SNP interactions on COPD risk. Multiple comparisons were performed using Bonferroni- correction.
ResultsOur results indicated that rs4719841 and rs7934083 variants were associated with a reduced risk of COPD. The analysis results of age, gender and non-smokers showed that rs4719841 and rs7934083 were associated with reducing the risk of COPD. In addition, the results showed that the genetic models of rs4719841, rs7934083 and rs7780562 were related to the forced vital capacity, respiratory rate per second, and respiratory rate / forced vital capacity of COPD patients, respectively. The results of the MDR analysis showed that the three-locus model (rs4719841, rs7934083, and rs78750958) is the best for COPD risk assessment.
ConclusionThis study shows that rs4719841 and rs7934083 are associated with the risk of COPD in the Chinese population, which provides some insights for early screening, prevention, and diagnosis of COPD in high-risk populations.
Chronic obstructive pulmonary disease (COPD) is a common, preventable, and treatable chronic lung disease that is one of the leading causes of death globally, killing more than 3 million people each year.1 It affects 329 million people worldwide, accounting for approximately 5% of the global population,2 imposes a huge economic burden on the health care system and has a significant social impact on patients and their families. Progress has been made in treating symptoms and preventing acute exacerbations; however little progress has been made in improving disease progression or affecting mortality. Therefore, a better understanding of the complex disease mechanisms that lead to COPD is urgently needed.3 In addition to smoking being a common cause of COPD, genetic factors play a significant role.4
Long-chain noncoding RNA molecules are approximately 200 nucleotides in length. They are well conserved, and defined as noncoding RNA molecules.5 However, some studies have shown that the peptide/protein encoded by ncRNAs.6 Lncrna is involved in many human diseases, such as prostate cancer,7 cardiovascular diseases8 and atherosclerosis.9 A ncRNA, Loc105375199 is located at 7p15.2, which is located on the short arm of human chromosome 7 and has band 2 in zone 15, with a total of nine exons. At present, there are few reports on Loc105375199; however, many studies have suggested that the gene on 8p22 may be involved in the occurrence and development of various lung diseases such as COPD.10–14 Although there are no detailed reports on COPD and Loc105375199, previous studies suggest that Loc105375199 may be involved in COPD occurrence. Therefore, it is necessary to study the relationship between the Loc105375199 polymorphism and the risk of COPD to provide a new perspective for preventing, screening, and treating COPD.
MIR4300HG is a non-coding RNA located on human chromosome 11. We found very few reports regarding MIR4300HGand most of them were related to spine diseases.15 In addition, many studies have shown that many genes on chromosome 11 are associated with COPD. T Ishii et al. revealed that polymorphism of the glutathione S-transferase P1 (GSTP1) gene located on chromosome 11 is related to COPD.16 Paul Stoll et al. found that patients with COPD had high levels of BDNF in their serum, which is encoded by the BDNF gene on chromosome 11.17 Multiple studies have shown that the cytokine IL18 located on chromosome 11 plays an important role in the inflammation in COPD.18–20 Therefore, we speculate that the MIR4300HG gene may be associated with the susceptibility to COPD.
To verify these speculations, we conducted a case-control study on the association between Loc105375199/MIR4300HG genetic polymorphisms and COPD susceptibility.
MethodsStudy participants and sample collectionThe project was approved by the medical ethics committee of the Chinese Medicine Hospital of Xinjiang Uygur Autonomous Region., All participants signed an informed consent form before the experiment. Data on environment, occupation, smoking history, tumors and general situation, according to the unified epidemiological questionnaire, was obtained from all patients. In this study, according to the GOLD (Global Chronic Obstructive Pulmonary Disease Action) report,21 315 clinical patients diagnosed with COPD were included in the case group, of which 147 were smokers (smoking more than two cigarettes per day, or had quit smoking before enrollment) and, 166 people were non-smokers (never smoked). The diagnosis of COPD by hospital pulmonologists is based on clinical examination and spirometry. The severity of COPD is classified according to fixed forced expiratory volume in 1 s (FEV1) / forced vital capacity (FVC <0.7) and the predicted value of FEV1. The healthy controls were 314 healthy subjects from the same geographic area with normal lung volume according to the GOLD standard, including 52 smokers and 118 non-smokers. Subjects with other lung diseases, infections, inflammation, endocrine, gastrointestinal, liver, kidney and tumor diseases, and excessive alcohol consumption (≥ 40 g /day) were excluded. Peripheral venous blood (5 ml) was collected in a test tube containing EDTA, centrifuged, and stored at - 80 ℃ for subsequent use.
SNP selection and genotypingTo verify the effectiveness of the genotyping of the selected population, the SNPs on chromosomes 11 and 7 were based on the smallest level according to the CHB (Beijing Han nationality) data in the Thousand Genome Project (http://www.internationalgenome.org/) SNPs with locus frequency less than 5% were initially screened. Subsequently, the HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was used to predict the potential functions of selected SNPs, which are related to promoter histone marks, enhancer histone marks, DNAse, proteins bound and selected eQTL hits, indicating the mechanisms by which these SNPs may affect the disease occurrence and development. Finally, we used a few previously reported SNPs located adjacent to LOC105375199 (rs10245353 and rs2290263), Loc105375199 (rs4719841 and rs7780562) and MIR4300HG (rs7934083 and rs78750958) as candidate loci for this study. Genomic DNA was extracted according to the GoldMag® nanoparticles method and its concentration was measured using the NanoDrop 2000. We conducted Multiplexed SNP MassEXTEND experiment, SNP genotyping and data management and analysis using the Sequenom MassARRAY Assay Design 3.0 software, Sequenom MassARRAY RS1000, and Sequenom Typer 4.0 software, respectively, as reported previously.22,23
Data analysisTo analyze the mass spectrometry results, the undetected, repeated data and the success rate of typing 90% of the sites or samples were deleted. This study used Welch's t-test to analyze the average age between the cases and control groups. Hardy Weinberg equilibrium (HWE) analysis is a method of quality control of the genetic data of the control group. Theoretically, the control group should follow the Hardy Weinberg equilibrium, p > 0.05, indicating that the control group reaches genetic balance, ensuring that the data of the control group is credible. Fisher precision test and Wald test were used to compare the allele/genotype frequency of each locus between the cases and the controls, and genetic model analysis was performed using SNPStats software (http://bioinfo.iconcologia.net/snpstats/start.htm) to assess the association between the SNPs and COPD risk. Multi-factor dimensionality reduction (MDR) analysis is a nonparametric, nongenetic approach suitable for case-control studies that only require individual genetic data (such as SNPs) and no other special conditions to analyze gene-gene interactions are needed.24,25 One-way analysis and ANOVA were used to analyze the differences in clinical characteristics between the different genetic models. Multiple comparisons were Bonferroni-corrected.26
ResultsStudy subjectsThe basic information of the participants in this study is shown in Table 1. A total of 629 subjects, 315 and 314 patients in the COPD and control groups, respectively, were included in this study. There were 239 men and 76 women in the COPD group and 237 men and 77 women in the control group. There were no significant differences in age, gender, smoking, complication, wheezing, chest tightness, recent respiratory infection status and BMI between the two groups. Detailed information such as the average value and standard deviation value of patients’ respiratory rate (per minute), pulse rate (pe minute), forced vital capacity (L), quantity per second (L), and rate per second (s) are shown in Table 1.
The basic information of the participants of the present study.
| Characteristics | Cases (N = 315) | Controls (N = 314) | p-value |
|---|---|---|---|
| Age | 0.306a | ||
| > 70years | 188(60%) | 177(56%) | |
| ≤ 70years | 127(40%) | 137(44%) | |
| Mean ±SD | 72.1 ± 8.5 | 69.9 ± 8.8 | |
| Gender | 0.908b | ||
| Male | 239(76%) | 237(75%) | |
| female | 76(24%) | 77(25%) | |
| Smoking status | |||
| yes | 147(47%) | 52(17%) | 0.057b |
| no | 166(53%) | 118(38%) | 0.156b |
| Complication status | 0.930b | ||
| yes | 93(30%) | 0 | |
| no | 174(55%) | 0 | |
| Wheezing status | 0.113b | ||
| yes | 153(49%) | 0 | |
| no | 123(39%) | 0 | |
| Gasp | < 0.001b | ||
| yes | 115(37%) | 0 | |
| no | 166(53%) | 0 | |
| Chest tightness status | 0.445b | ||
| yes | 102(32%) | 0 | |
| no | 179(57%) | 0 | |
| Recent respiratory infection status | 0.788b | ||
| yes | 164(52%) | 0 | |
| no | 117(37%) | 0 | |
| BMI | 0.015a | ||
| >24 | 251(80%) | 67(21%) | |
| ≤24 | 29(9%) | 78(25%) | |
| Respiratory rate (per minute) | 22.31±2.462 | ||
| Pulse rate (per inute) | 86.33±11.675 | ||
| Forced vital capacity (L) | 2.26±3.717 | ||
| Quantity per second (L) | 1.22±0.600 | ||
| Rate per second (s) | 39.91±29.329 | ||
| GOLD spirometric grade, n (%) | |||
| Ⅰ | 34(10.8%) | ||
| Ⅱ | 147(46.7%) | ||
| Ⅲ | 102(32.4%) | ||
| Ⅳ | 32(10.2%) |
SD: Standard deviation; COPD:chronic obstructive pulmonary disease; BMI: Body mass index.
p < 0.05 indicates statistical significanc..
The basic information of all the SNPs is shown in Table 2, including SNP-ID, gene, chromosomes, position, alleles, minor allele frequency (MAF), p-HWE, OR (95%CI) and p-value. After Bonferroni correction, the results showed that individuals with the A allele of rs4719841 had a lower risk of COPD than those with the G allele (p < 0.0001). Individuals with the G allele of rs78750958 had a lower risk of COPD than those with the A allele (p < 0.0001). HaploReg software function prediction showed that the selected SNPs (rs10245353 rs2290263, rs4719841, rs7780562, rs7934083 and rs78750958) are related to siPhy cons, DNAse, motif changes, selected eQTL hits, promoter histone marks, enhancer histone marks, and proteins, suggesting that these SNPs may affect the occurrence and development of the disease. A multi-genetic model was used to analyze the correlation between the selected SNPs and the risk of COPD, and the results showed that rs4719841 and rs7934083 were related to the risk of COPD (p < 0.0017) (Table 3). Specifically, the A/A genotype of rs4719841 was observed to reduce the risk of COPD. In the co-dominant, dominant and additive models, rs7934083 reduced the risk of COPD.
Basic information of the selected SNPs in this study.
SNP: Single nucleotide polymorphism; HWE: Hardy-Weinberg equilibrium; OR: Odds ratio; 95% CI: 95% confidence interval; COPD:chronic obstructive pulmonary disease.HWE p-value obtained from Chi-squared test.
p values were calculated from Chi-squared test regarding to the allele distribution frequencies among COPD and healthy controls.
Bonferroniʼs multiple adjustment was applied, with p < 0.0017(0.05/30).
Bold indicates a statistically significant SNP (p < 0.0017).
Relationships of the SNPs and the risk of COPD.
SNP: Single nucleotide polymorphism; OR: Odds ratio; 95% CI: 95% confidence interval; COPD:chronic obstructive pulmonary disease.
p-values were calculated by unconditional logistic regression analysis with the comparison between COPD and healthy controls with adjustments for age and gender.
Bonferroniʼs multiple adjustment was applied, with p < 0.0017(0.05/30).
Bold indicates a statistically significant SNP (p < 0.0017).
To further clarify the correlation between the six selected SNPs and the risk of COPD, we conducted 10 stratified analyses, including stratification by age, gender, complications, BMI, asthma, recent respiratory infection, smoking, wheezing, chest tightness, and GOLD spirometric grade (Supplementary Table 1). The results showed that COPD risk due to some of the selected SNPs was associated with age, gender, and non-smoking categories, whereas in case of other selected SNPs, COPD risk was not associated with stratified analyses.
In subjects aged > 70 years, we found that rs7934083 was associated with reducing COPD risk (p < 0.0017), while in subjects aged ≤ 70 years, rs4719841 was associated with a reduction in the risk of COPD (p < 0.0017) (Table 4). The results of the male stratification analysis showed that rs4719841 reduced the risk of COPD in the allele, collinearity model, additive model and recessive model (p < 0.0017). The results of the male stratification analysis also showed that individuals with the T allele rs7934083 had a lower risk of COPD than individuals with the C allele (p = 0.0007). In the female stratification analysis, we only observed that rs7934083 was associated with a reduced risk of COPD (p < 0.0017). In the stratified analysis of non-smokers, we observed that the other five selected SNPs were not related to the risk of COPD except for rs4719841, which is related to a reduced risk of COPD (Table 5).
Stratified analysis of the age and gender on association between the SNPs and COPD risk.
SNP: single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; COPD:chronic obstructive pulmonary disease.
Bonferroniʼs multiple adjustment was applied, with p < 0.0017(0.05/30).
Bold indicates a statistically significant SNP (p < 0.0017).
Stratified analysis of the smoking on association between the SNPs and COPD risk.
SNP: single-nucleotide polymorphism; OR, odds ratio; CI, confidence interval; COPD:chronic obstructive pulmonary disease.
Bonferroniʼs multiple adjustment was applied, with p < 0.0017(0.05/30).
Bold indicates a statistically significant SNP (p < 0.0017).
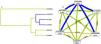
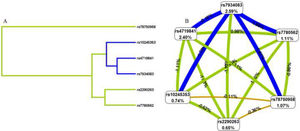
To better understand the impact of selected SNP interactions on COPD risk, we conducted MDR analysis (Supplementary Table 2 and Fig. 1). The dendrogram and frauctman-reiringold represent the interactions between these SNPs. In Fig. 1(A), the connection between nodes is shorter, indicating that the redundant interaction is stronger. A negative double-track entropy in Fig. 1(B) indicates that it is an antagonistic effect. A positive value indicates a synergistic effect. The results showed that there was a specific antagonism between the selected genes. Supplementary Table 2 shows the model combination of candidate SNP interactions for the COPD risk assessment. Among the many locus models, the best combination is a three-locus model consisting of rs4719841, rs7934083 and rs78750958, which increases the risk of COPD (test accuracy=0.618, CVC=10/10, OR=2.93, 95% CI: 2.11 – 4.05, p < 0.001).3.4 3.5 Genetic models and clinical characteristics.
In addition, we explored the relationship between different SNP models and the clinical indices of patients, including respiratory rate (times per minute), pulse rate (times per minute), forced vital capacity (L), one-second volume (L), and one- second rate (S). There was no significant correlation between the genetic models of rs10245353, rs2290263, and rs78750958 and the above clinical parameters (p > 0.05). However, the genetic models of rs4719841, rs7934083, and rs7780562 are related to forced vital capacity, rate per second, and respiratory rate/forced vital capacity of COPD patients (p < 0.05), as shown in Table 6.
Clinical characteristics of patients based on the genetic models of the SNPs.
SNP: single-nucleotide polymorphism; COPD: :chronic obstructive pulmonary disease; “-” indicates data missing;.
Bonferroniʼs multiple adjustment was applied, with p < 0.05.
Bold indicates a statistically significant SNP (p < 0.05).
Based on a case-control study, we investigated the relationship between the six SNPs (rs10245353, rs2290263, rs78750958, rs4719841, rs7934083, and rs7780562) and susceptibility to COPD in the Chinese population. In the overall analysis, only two SNPs (rs4719841, rs7934083) were found to be associated with susceptibility to COPD. In multiple stratified analyses, we observed that only some selected SNPs were associated with COPD risk in the stratified analysis of age, gender, and non-smoking. The results of the MDR analysis showed that the three-locus model comprising rs4719841, rs7934083 and rs78750958 is the best model for COPD risk assessment. In the analysis of clinical indicators, the genetic models of rs4719841, rs7934083, and rs7780562 were associated with forced vital capacity, respiratory rate per second, and respiratory rate/forced vital capacity, respectively.
COPD is chronic bronchitis and emphysema with airflow obstruction characteristics. It is currently one of the leading causes of chronic morbidity and mortality.27 Studies have shown that genetic factors play a key role in the occurrence and development of COPD.28 Genetic association research is an important way to understand the pathogenesis and epidemiology of COPD. A study by Almira Akparova et al. showed that the EPHX1 Y113H polymorphism is associated with an increased risk of COPD in the Kazakhstan population.29 Studies have also shown that vitamin D binding protein (VDBP), promoter regions of matrix metalloproteinases, cyclooxygenase-2 (COX-2), and angiotensin converting enzyme gene (ACE) polymorphisms are associated with COPD susceptibility.30–33
Non-coding RNAs are associated with various diseases.7-9 However, even searching many databases, no report of Loc105375199, a non-coding RNA, was found. Our study showed that the polymorphism of Loc105375199 is associated with susceptibility to COPD; that is, rs4719841 in the Loc105375199 gene is associated with a reduction in susceptibility to COPD. This is the first report on Loc105375199 and COPD, which provides a unique perspective for future studies on Loc105375199 and COPD and a new direction for t follow-up studies.
A previous study only involved rs10245353, located at the locus adjacent to Loc105375199, which indicated that rs10245353 mutation may increase the risk of gestational diabetes mellitus (GDM) in the Chinese population.34 There was no statistically significant relationship between rs10245353 mutation and COPD risk in our results, which may also be because our sample size was not large enough; this should be verified by further expanding sample size and mechanism studies.
MIR4300HG is a host gene for miRNAs. There is only one report on MIR4300HG, indicating that the functional variation of MIR4300HG is related to the progression of adolescent idiopathic scoliosis (AIS).15 Our study showed that rs7934083 on MIR4300HG was associated with a reduced risk of COPD, which also verified our hypothesis.
This study has several limitations. First, the sample size of this study was not large enough. Therefore, we still need to expand the sample size to verify the results further. Second, this study only involved related studies, and cell and animal experiments are needed to clarify the specific mechanism further.
ConclusionThis study is the first to report that the polymorphisms of Loc105375199 and MIR4300HG are associated with COPD susceptibility, which provides a new direction for the study of COPD and is more conducive to the promotion of personalized medicine and pharmacogenomics.