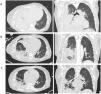
A 62-year-old female patient was admitted to the emergency department (ED) with a 7-day presentation of worsening cough, dyspnea, and pleuritic chest pain. She was unemployed and past medical history included a follicular lymphoma in remission and ischemic heart disease. She was vaccinated against SARS-CoV-2 and Influenza. Upon admission, the patient was hemodynamically stable, afebrile, tachypneic, with a peripheral oximetry of 93 % on oxygen via nasal cannula (1 L/min). Pulmonary auscultation of the right hemithorax was reduced. Arterial blood gas demonstrated adequate oxygenation and respiratory alkalosis. Laboratory evaluation revealed neutrophilic leukocytosis and high C-reactive protein. SARS-CoV-2 antigen and polymerase chain reaction were positive (cycle threshold [Ct] 29.4). Computerized tomography (CT) showed lung consolidation of the left superior and inferior lobes with a loculated pleural effusion (Fig. 1A). Empirical therapy with ceftriaxone and clarithromycin was started.
A. Admission lung CT: extensive lung consolidation of the superior and inferior right lobes and large loculated pleural effusion; B. Day 7 lung CT (after chest drainage insertion):large heterogeneous lung consolidation with air bronchogram and gas-filled cavities predominantly in the apical and posterior basal segments of the lower right lobe; C. Day 22 lung CT (after chest drainage repositioning and removal): improvement of lung consolidation extension and pleural effusion, with 16 days of linezolid.
The patient deteriorated over 48 h with severe hypoxemic respiratory failure, requiring endotracheal intubation and ICU admission. Thoracocentesis revealed empyema that was drained with improvement of lung mechanics. Positive urinary antigen for Streptococcus pneumoniae motivated seven days of ceftriaxone and three days of clarithromycin. Microbiological testing showed MSSA on bronchoalveolar lavage (BAL) and tracheal aspirate, with a positive respiratory viral panel for rhinovirus. Complete microbiological workup is described in Table 1. On the seventh day of admission, thoracic CT showed signs of necrotizing pneumonia with no evidence of obliteration in the arterial supply, such as segmental thrombosis (Fig. 1B). The possibility of a PVL-producing MSSA strain was considered, and antimicrobial therapy was altered to linezolid. Bacterial phenotype and genotype were requested. The MSSA clonal lineage was determined based on the sequencing of an internal fragment of the spa gene. MSSA isolate was identified as belonging to spa type t1451, commonly associated with clonal complex CC398. It showed resistance to clindamycin, having a susceptible phenotype to other antibiotics (n = 21), including cefoxitin. The PVL gene was not detected, however the genes encoding β- and γ-hemolysin were. The chp and scn genes of the Immune Evasion Cluster (IEC) were found through PCR testing, suggesting an underlying association with the CC398 human clade. The patient was extubated after 10 days of invasive mechanical ventilation. Pulmonary lobectomy and decortication were deemed unnecessary upon improvement of lung consolidation and pleural effusion (Fig. 1C). The patient was transferred to an intermediate-care unit and antibiotics maintained for five weeks, despite the switch to flucloxacillin. She was subsequently transferred to the Pulmonology ward where she underwent rehabilitation and weaning from oxygen therapy for one month. After that she was discharged to a rehabilitation unit and return home after three months, autonomous, despite maintaining some tiredness, and without the need for supplemental oxygen.
Microbiological workup in ICU.
| Microbiological Test | Specimen | Result | Timing of ICU stay (days) |
|---|---|---|---|
| Bacterial cultures | Tracheal aspirate | Negative | 1 |
| Pleural effusion (exudate/empyema) | Negative | 2 | |
| Blood culture | Negative | 2 | |
| BAL | MSSA | 4 | |
| Tracheal aspirate | MSSA | 4 | |
| Blood culture | Negative | 6 | |
| CVC | Negative | 6 | |
| Arterial line | Negative | 6 | |
| Tracheal aspirate | Negative | 9 | |
| BAL | Negative | 9 | |
| Pleural effusion | Negative | 9 | |
| BAL (FB)ª | Negative | 15 | |
| Other Microbiological Tests | |||
| Streptococcus pneumoniae antigen | Urine | Positive | 1 |
| Legionella pneumophila antigen | Urine | Negative | 1 |
| SARS-CoV-2 PCR | Nasopharyngeal swab | Positive (high CT)* | 1 |
| SARS-CoV-2 antigen | Nasopharyngeal swab | Positive | 1 |
| MRSA PCR⁎⁎ | Nasal swab | Negative | 1 |
| Respiratory virus panel | Nasopharyngeal swab | Positive:- Rhinovirus- SARS-CoV-2 | 1 |
| Ac Anti-HIV 1/2 e Ag P24 HIV 1 (chemiluminescence test) | Plasma | Negative | 1 |
| Mycobacterium stain (Ziehl Neelsen) | Pleural effusion | Negative | 2 |
| Galactomannan test | BAL⁎⁎⁎ | Negative | 4 |
| Streptococcus pneumoniae antigen | BAL | Negative | 4 |
| Legionella spp. (culture) | BAL | Negative | 4 |
| Mycological culture | BAL | Negative | 4 |
| Mycobacterium stain (Ziehl Neelsen) | BAL | Negative | 4 |
| Mycobacterium culture | BAL | Negative | 4 |
| Mycological culture | Pleural effusion | Negative | 9 |
| Mycobacterium stain (Ziehl Neelsen) | Pleural effusion | Negative | 9 |
| Mycobacterium culture | Pleural effusion | Negative | 9 |
| HSV-1/HSV-2 PCR | Oral mucosa/vesicles | Positive/Negative | 11 |
| varicella-zoster virus PCR | Oral mucosa/vesicles | Negative | 11 |
| MRSA PCR | Nasal swab | Negative | 29 |
ªFB - Flexible bronchoscopy.
This is the first case of severe necrotizing community-acquired pneumonia due to a non-PVL MSSA CC398 strain reported in Portugal. This documentation is important since this agent, belonging to the human clade, has been frequently associated with colonization.1
Although the patient might have initially presented with a community-acquired pneumococcal infection, the clinical course of necrotizing pneumonia was attributed to MSSA. S.pneumoniae was never isolated in cultures and urinary antigens can persist after acute infection. The positive results for rhinovirus and SARS-CoV-2 could have influenced the clinical severity of the case. The interactions between viral and bacterial infections have been widely studied. However, the simultaneous positivity of rhinovirus PCR on the respiratory panel does not implicate it as an acute infectious agent, but it is frequently identified alongside S.aureus. After infection clearance, respective viral loads decrease.2 SARS-CoV-2 pneumonia may coexist or even trigger a bacterial pneumonia.3 However, in the case of our patient, the role of SARS-CoV-2 in determining the clinical-radiological aspects is not relevant. She presented high Ct and the dominant clinical picture of unilateral pneumonia with empyema favor a bacterial agent.
Necrotizing pneumonia caused by MSSA CC398 was rarely reported in the past. The most common clinical presentations of invasive disease were osteomyelitis and bloodstream infections.4 What might be considered an easily treatable MSSA strain can, in fact, be a more virulent agent with implications regarding antibiotic choices. In our case, linezolid was essential due to its favorable pharmacokinetic properties (better lung penetration). Although the MSSA strain was associated with a CC398 clone, the patient did not have a history of contact with either livestock or other animals. Furthermore, the isolate was PVL-negative and susceptible to tetracycline and methicillin, possessing genes typically carried within the IEC. This is in accordance with recent documentation of MSSA CC398 infections in young patients without previous animal contact.5 The virulence mechanisms of CC398 MSSA human clade are unclear. Recent studies suggest that MSSA CC398 are becoming more resistant to antibiotics, more pathogenic to humans and have gained the ability to spread in the community and hospital settings.6 This has been attributed to the acquisition of bacteriophages, leading to increased immune evasion and higher adhesion and invasion capacity of epithelial cells.7 Different levels in the expression of α-toxin, phenol-soluble modulin and protein A, were also detected between ST398 and non-ST398 pneumonia isolates, possibly contributing to virulence [28]. It was shown that ST398 had an increased capacity to lyse human erythrocytes and neutrophils and cause severe multifocal necrotizing pneumonia,7 as observed in this case. In cases of such clinical severity, we recommend the genotypic characterization, motivating further testing and active discussion with Microbiology Units to better classify underlying agents and strains that would otherwise persist unidentified.
Diana Caieirob – ORCID 0000-0001-8637-6778; d.pinho@campus.fct.unl.pt, Hermínia de Lencastrec,d – ORCID 0000-0001-6816-8932; Maria Miragaiab – ORCID 0000-0002-1323-7184; miragaia@itqb.unl.pt; Pedro Póvoa a,e,f – ORCID 0000-0002-7069-7304; pedrorpovoa@gmail.com a Department of Intensive Care (ICU4), Hospital de São Francisco Xavier – Centro Hospitalar Lisboa Ocidental, Estrada do Forte do Alto Duque, 1449-0054 Lisbon, Portugal, b Laboratory of Bacterial Evolution and Molecular Epidemiology, Instituto de Tecnologia Química e Biológica António Xavier (ITQB-NOVA), Universidade Nova de Lisboa, Avenida da República, 2780-157 Oeiras, Portugal, c Laboratory of Molecular Genetics, Instituto de Tecnologia Química e Biológica António Xavier (ITQB-NOVA), Universidade Nova de Lisboa, Avenida da República, 2780-157 Oeiras, Portugal, d Laboratory of Microbiology & Infectious Diseases, The Rockefeller University, 1230 York Av, New York, NY10065, USA, eNova Medical School – New University of Lisbon, Campo dos Mártires da Pátria 130, 1169-056 Lisbon, Portugal, f Center of Clinical Epidemiology and Research Unit of Clinical Epidemiology, OHU Odense University Hospital, 5000 Odense, Denmark