Lung cancer has the highest incidence and mortality worldwide.1 Nowadays target therapy is the first line therapy in metastatic non-small cell lung cancer (NSCLC) in patients with target mutations.2,3 Next-generation sequencing (NGS) permits simultaneous reading of DNA sequences, with deep sequencing, allowing for the detection of genetic variants that occur at low percentages,4 in a short period of time and with relatively low cost.5
The aim of this study was to evaluate the mutational profile in patients with NSCLC, diagnosed in a Tertiary Hospital, which performed molecular testing by NGS.
We carried out a retrospective review of 204 patients with NSCLC who performed NGS in the Thoracic Tumors Multidisciplinary Unit of Vila Nova de Gaia-Espinho Hospital Center between April 2016 and May 2018. We included in our study only patients with at least one mutation identified by NGS. Sociodemographic and clinical data were retrospectively reviewed from clinical files. Descriptive statistics were used to analyze patient's characteristics.
Our NGS technique include the “Oncomine Solid Tumor DNA” and the “Oncomine Solid Tumor Fusion Transcript” panels that allow for the identification of variants in the genes EGFR, KRAS, NRAS, BRAF, MET, ERBB4 (HER4), ERBB2 (HER2), ALK, PI3KCA and PTEN and gene rearrangements in ALK, ROS1 and RET. This NGS technique, developed and validated by IPATIMUP Diagnostics, allowed us, with a sensibility >99%, to detect nucleotide substitutions with allelic fraction >5% and rearrangements in 1% of the RNA, in samples with more than 20% of neoplastic cells.
As a result, of the 204 patients with NSCLC who performed NGS 121 (59%) had some mutation and therefore were included. Patients were mostly male (n=71; 58.7%), with a mean age of 66±10 years-old. Most patients were smokers (n=36; 30%) or former smokers (n=44; 36%) and had a performance status <2 (n=103; 85%). NGS was performed prior to any line of treatment in 109 patients (90%). For the remaining 10% of patients (n=12) NGS was performed in the course of the disease, after one or more lines of treatment.
Adenocarcinoma was the most frequent histological type (n=110; 90.9%). Others histological types were: squamous cell carcinoma (n=5; 4.1%), NSCLC not otherwise specified (n=5; 4.1%) and large cell neuroendocrine lung carcinoma (n=1; 0.8%).
According to TNM Staging, most patients were in stage IVB (n=34; 28.1%) or stage IVA (n=33; 27.3%) when NGS was performed.
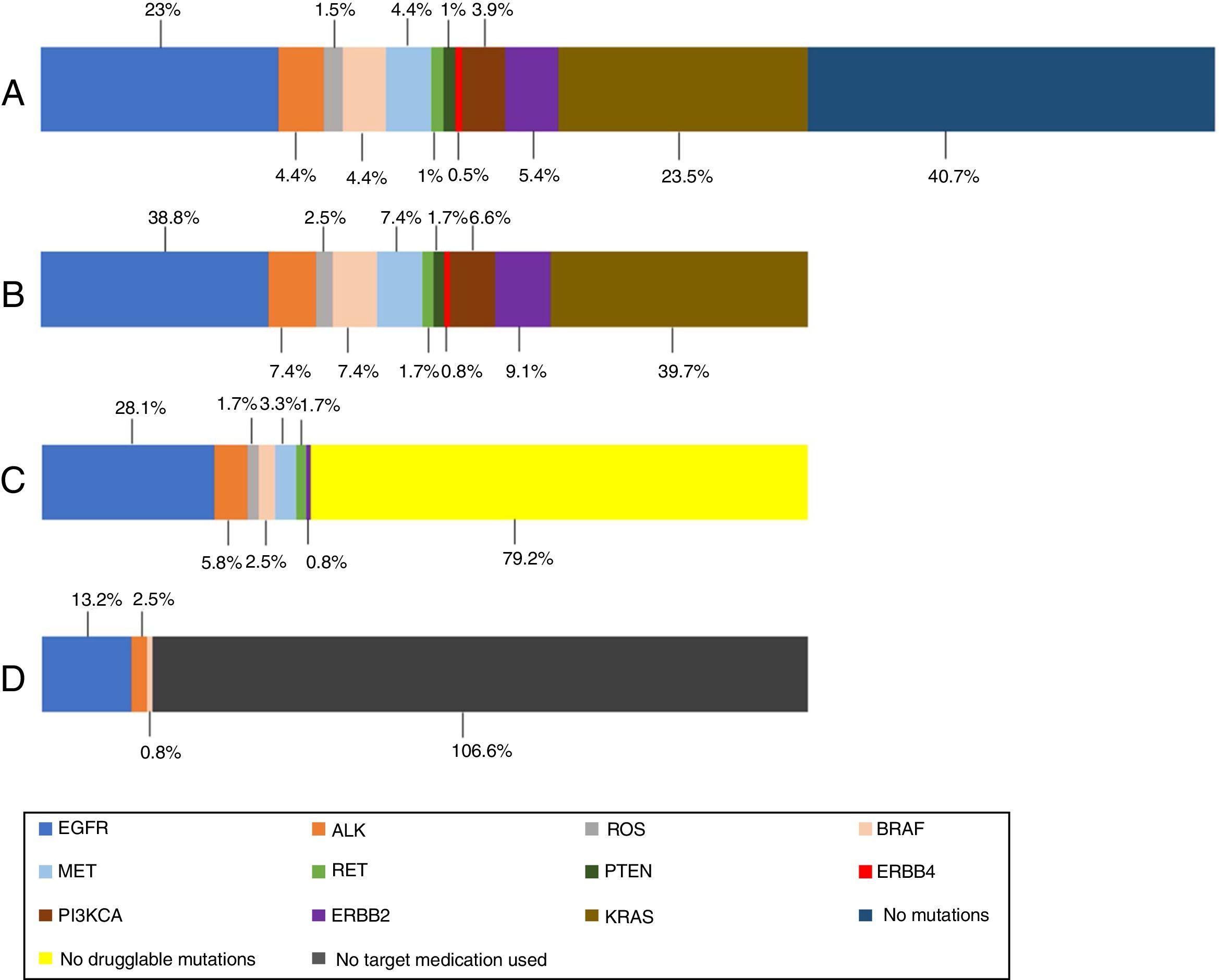
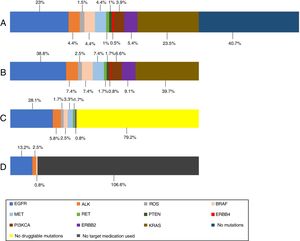
In the 121 patients, 149 mutations were identified (Fig. 1). The genes more frequently mutated were KRAS (n=48) and EGFR (n=47). Overall prevalence of each mutation in patients who performed NGS are also presented in Fig. 1.
(A) Prevalence of mutations in all NGS patients; (B) prevalence of mutations in NGS positive patients; (C) prevalence of druggable mutations in NGS positive patients (off-label regimen in case of MET, RET and ERBB2 mutations); (D) prevalence of treated mutations in NGS positive patients. KRAS (n=48); EGFR (n=47); ERBB2 (n=11); MET (n=9); BRAF (n=9); ALK (n=9); PI3KCA (n=8); ROS (n=3); RET (n=2); PTEN (n=2); ERBB4 (n=1).
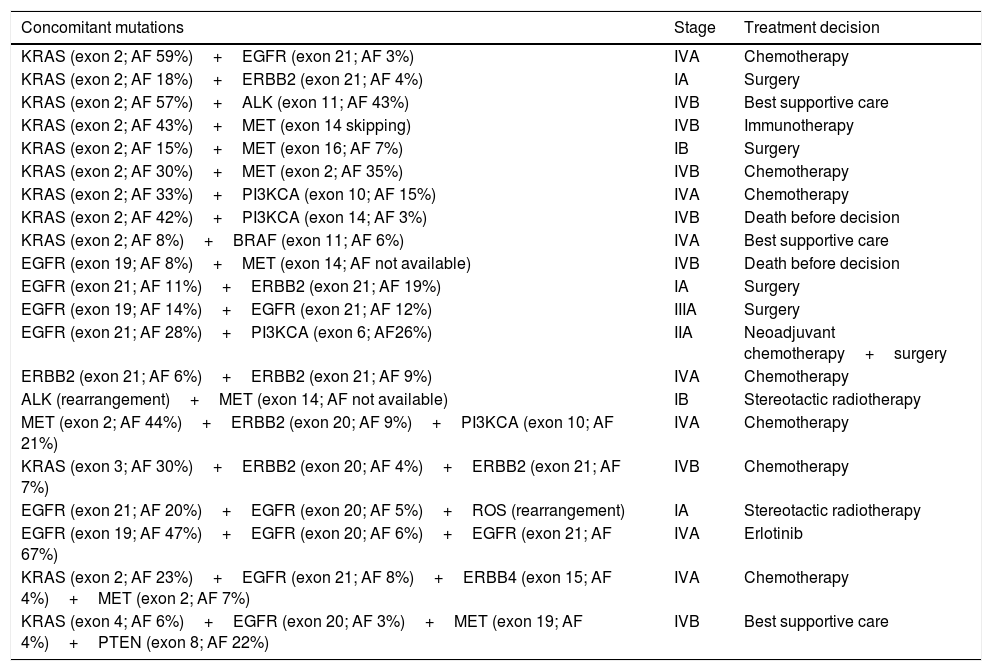
More than one mutation was identified in 21 patients (17.3%): 4 patients had 3 concomitant mutations and 2 patients had 4 concomitant mutations (Table 1). Of those patients, just one single case was treated with target therapy, after analysis of the mutational profile and the allelic frequencies of each mutation. For this case, the tyrosine kinase inhibitor was chosen based on the two mutations with higher allelic frequencies.
Concomitant mutations and treatment decision.
| Concomitant mutations | Stage | Treatment decision |
|---|---|---|
| KRAS (exon 2; AF 59%)+EGFR (exon 21; AF 3%) | IVA | Chemotherapy |
| KRAS (exon 2; AF 18%)+ERBB2 (exon 21; AF 4%) | IA | Surgery |
| KRAS (exon 2; AF 57%)+ALK (exon 11; AF 43%) | IVB | Best supportive care |
| KRAS (exon 2; AF 43%)+MET (exon 14 skipping) | IVB | Immunotherapy |
| KRAS (exon 2; AF 15%)+MET (exon 16; AF 7%) | IB | Surgery |
| KRAS (exon 2; AF 30%)+MET (exon 2; AF 35%) | IVB | Chemotherapy |
| KRAS (exon 2; AF 33%)+PI3KCA (exon 10; AF 15%) | IVA | Chemotherapy |
| KRAS (exon 2; AF 42%)+PI3KCA (exon 14; AF 3%) | IVB | Death before decision |
| KRAS (exon 2; AF 8%)+BRAF (exon 11; AF 6%) | IVA | Best supportive care |
| EGFR (exon 19; AF 8%)+MET (exon 14; AF not available) | IVB | Death before decision |
| EGFR (exon 21; AF 11%)+ERBB2 (exon 21; AF 19%) | IA | Surgery |
| EGFR (exon 19; AF 14%)+EGFR (exon 21; AF 12%) | IIIA | Surgery |
| EGFR (exon 21; AF 28%)+PI3KCA (exon 6; AF26%) | IIA | Neoadjuvant chemotherapy+surgery |
| ERBB2 (exon 21; AF 6%)+ERBB2 (exon 21; AF 9%) | IVA | Chemotherapy |
| ALK (rearrangement)+MET (exon 14; AF not available) | IB | Stereotactic radiotherapy |
| MET (exon 2; AF 44%)+ERBB2 (exon 20; AF 9%)+PI3KCA (exon 10; AF 21%) | IVA | Chemotherapy |
| KRAS (exon 3; AF 30%)+ERBB2 (exon 20; AF 4%)+ERBB2 (exon 21; AF 7%) | IVB | Chemotherapy |
| EGFR (exon 21; AF 20%)+EGFR (exon 20; AF 5%)+ROS (rearrangement) | IA | Stereotactic radiotherapy |
| EGFR (exon 19; AF 47%)+EGFR (exon 20; AF 6%)+EGFR (exon 21; AF 67%) | IVA | Erlotinib |
| KRAS (exon 2; AF 23%)+EGFR (exon 21; AF 8%)+ERBB4 (exon 15; AF 4%)+MET (exon 2; AF 7%) | IVA | Chemotherapy |
| KRAS (exon 4; AF 6%)+EGFR (exon 20; AF 3%)+MET (exon 19; AF 4%)+PTEN (exon 8; AF 22%) | IVB | Best supportive care |
AF: allelic frequency.
Target therapy, as first line treatment, was started in 30% of stage IV patients (n=20): erlotinib (n=8), gefitinib (n=3), afatinib (n=3), crizotinib (n=2), erlotinib+bevacizumab (n=2), alectinib (n=1) and dabrafenib+trametinib (n=1).
Target therapy has become the first line of treatment in patients with metastatic NSCLC with target mutations. Until a few years ago, single gene analysis methods were the main option for performing mutational profile analysis. However, this technique is time consuming and requires a large amount of tumor DNA, which may not be available. NGS has become an alternative because it enables a simultaneous multiple gene analysis, using less DNA and is a quick and cost-effective technique. In the present study, 59% of patients that performed NGS presented a molecular variant in the genes of our NGS panel which made allowed us to start first line target therapy in 30% of stage IV patients.
In our study, many tumors had more than one mutation, with 6 patients having more than 3 concomitant mutations, which is rarely described in the literature. More than one patient had 2 or 3 mutations identified in the same gene and there were patients with more than one mutation suitable for target therapy. This was detected because sensitivity obtained by NGS is superior to the previous molecular techniques, making possible the detection of genetic variants that occur at low percentages. This mutational profile with concomitant mutations probably reflects tumor heterogeneity and can help us to personalize the therapeutics but, at same time, makes the therapeutics decisions more difficult in patients with more than one mutation.
Our study is the first presentation of NGS data in Portugal. Since is a retrospective single center study, the sample is relatively small. It might be interesting to create a multicenter database to provide a registration of the Portuguese NSCLC mutational profile.
In conclusion, the use of NGS has been increasing and has allowed us to evaluate more accurately the mutational profile of the tumors and detect genetic variants that occur at low percentages. This allows for the use of already approved target therapy or the integration of patients into clinical trials. On the other hand, the detection of concomitant mutations makes the therapeutic decisions in patients with NSCLC even more complex and challenging.
FundingThis research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interestNone to declare.